Figure 1.
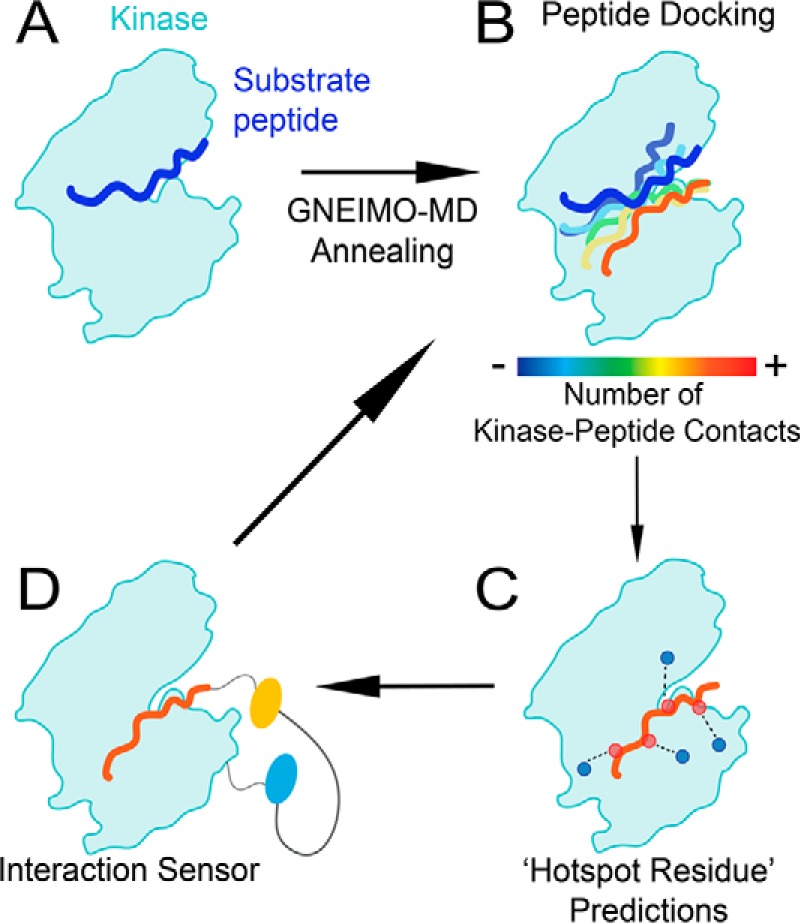
Combining computational and experimental approaches to dissect the kinase–substrate interaction. A, PKC–substrate interaction is homology modeled from the PKCι–Par3 interaction (PDB ID 5HI1). B, GNEIMO torsional MD is used to obtain a kinase–substrate conformational ensemble. C, structural analysis is used to identify hot spot residues and motifs within the kinase and substrate that contribute to binding and catalytic activity. D, SPASM FRET sensors are used to test GNEIMO-MD predictions using site-directed mutagenesis. Experimental results are used to validate GNEIMO-MD predictions and refine computational methods.
