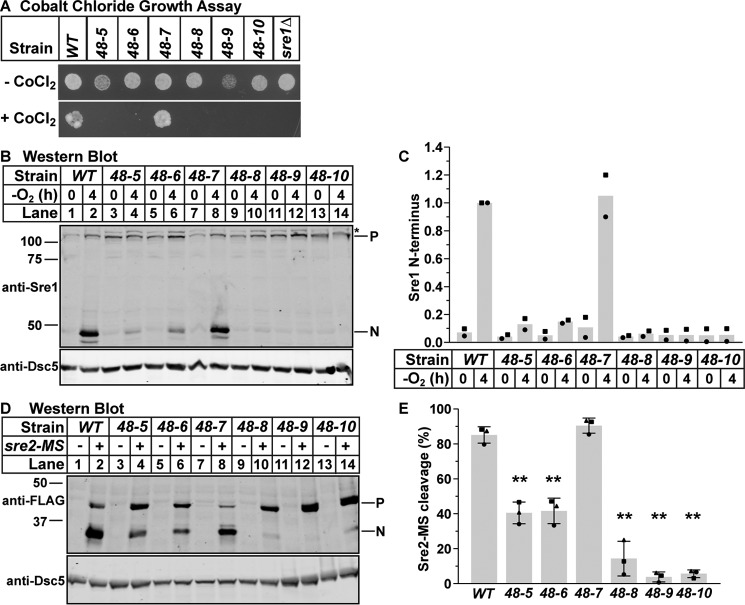
Figure 2.
SREBP cleavage requires cdc48. A, wild-type cells or the indicated mutants (5000 cells) were grown on rich medium plus or minus CoCl2 for 2 or 10 days, respectively. B, Western blots, probed with monoclonal anti-Sre1 IgG (5B4) and polyclonal anti-Dsc5 IgG (for loading), of lysates treated with alkaline phosphatase for 1 h from wild-type cells and the indicated cdc48 mutants grown for 0 or 4 h in the absence of oxygen. P and N denote precursor and cleaved N-terminal transcription factor forms, respectively. Asterisk denotes non-specific band. C, quantification of Sre1 N terminus from B of two biological replicates each denoted by different marker symbols. The quantity of Sre1N was normalized to Dsc5 for loading and then to the WT 4-h sample (lane 2) for comparison between blots. D, Western blots, probed with monoclonal anti-FLAG M2 and polyclonal anti-Dsc5 IgG (for loading), of lysates treated with alkaline phosphatase for 1 h from wild-type cells and the indicated cdc48 mutants containing a plasmid expressing sre2-MS (+) or the empty vector (−) grown in the presence of oxygen. P and N denote precursor and cleaved N-terminal transcription factor forms, respectively. E, quantification of Sre2-MS cleavage from D of three biological replicates each denoted by different marker symbols. The quantity of precursor and N terminus were normalized first to empty vector and then to Dsc5 for loading. Percent cleavage in each sample was calculated by dividing the normalized quantity of N terminus by the total signal (N+P). Error bars are 1 S.D. (**, p < 0.01 versus WT by two-tailed Student's t test).