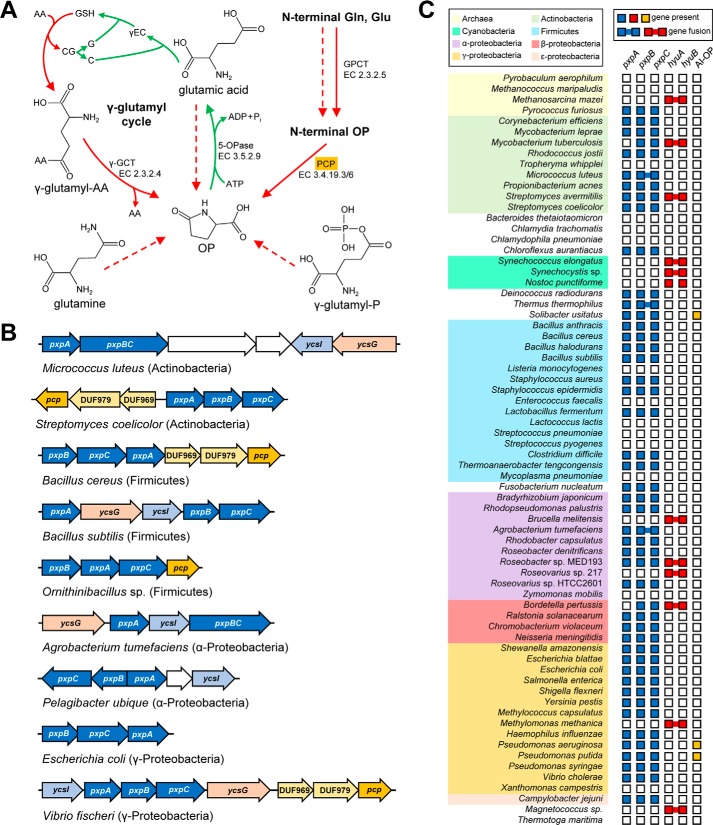
Figure 1.
OP metabolism and the genomic organization and distribution of OP metabolic genes in prokaryotes. A, OP is formed spontaneously (dashed red arrows) and/or enzymatically (solid red arrows) from glutamine, glutamate, γ-glutamyl phosphate (γ-glutamyl-P), N-terminal glutaminyl or glutamyl residues of peptides (GPCT, glutaminyl-peptide cyclotransferase), and γ-glutamyl-amino acids (γ-glutamyl-AA) via the γ-glutamyl cycle (indicated with curved arrows; AA, amino acid; CG, cysteinyl-glycine; C, cysteine; G, glycine; γEC, γ-glutamyl-cysteine; γ-GCT, γ-glutamylcyclotransferase). Hydrolysis to glutamate is the only way to recycle OP. Green arrows indicate glutathione synthesis reactions of the γ-glutamyl cycle. B, clustering and fusion of the candidate pxpABC OP metabolism genes in representative genomes. The pxpABC genes (colored blue) often cluster with genes encoding pyroglutamyl peptidase (PCP; colored orange), two conserved membrane-spanning proteins (DUF969 and DUF979; colored yellow), an NRAMP family transporter (ycsG; colored pink), and a putative metal chaperone (ycsI; colored light blue). Genes apparently unrelated to OP metabolism are colored white. C, distribution of candidate pxpABC OP metabolism genes and known 5-oxoprolinase genes among a representative set of 70 diverse bacteria and archaea. hyuA and hyuB represent eukaryotic-type 5-oxoprolinase homologs; AI-OP represents ATP-independent 5-oxoprolinase homologs.