Abstract
Although the signal pathways mediating muscle protein synthesis and degradation are well characterized, the transcriptional processes modulating skeletal muscle mass and adaptive growth are poorly understood. Recently, studies in mouse models of muscle wasting or acutely exercised human muscle have suggested a potential role for the transcription factor signal transducer and activator of transcription 3 (STAT3), in adaptive growth. Hence, in the present study we sought to define the contribution of STAT3 to skeletal muscle adaptive growth. In contrast to previous work, two different resistance exercise protocols did not change STAT3 phosphorylation in human skeletal muscle. To directly address the role of STAT3 in load-induced (i.e., adaptive) growth, we studied the anabolic effects of 14 days of synergist ablation (SA) in skeletal muscle-specific STAT3 knockout (mKO) mice and their floxed, wild-type (WT) littermates. Plantaris muscle weight and fiber area in the nonoperated leg (control; CON) was comparable between genotypes. As expected, SA significantly increased plantaris weight, muscle fiber cross-sectional area, and anabolic signaling in WT mice, although interestingly, this induction was not impaired in STAT3 mKO mice. Collectively, these data demonstrate that STAT3 is not required for overload-mediated hypertrophy in mouse skeletal muscle.
Keywords: STAT3, skeletal muscle, hypertrophy, resistance exercise
skeletal muscle wasting is a hallmark of several diseases, including cancer cachexia, chronic kidney disease (CKD), heart failure, and diabetes (4, 14). These diseases are characterized by elevated levels of circulatory proinflammatory cytokines, which subsequently promote skeletal muscle atrophy (4, 14). Resistance exercise and dietary proteins are key stimuli to offset skeletal muscle wasting, mainly by increasing the rate of protein synthesis via phosphoinositide 3-kinase (PI3K) and mechanistic target of rapamycin complex 1 (mTORC1) pathways (7, 10). Although the transcriptional mechanisms regulating resistance exercise-mediated skeletal muscle hypertrophy are largely unknown, acute resistance exercise has been reported to increases the activity of the transcription factor signal transducer and activator of transcription 3 (STAT3) in human (5, 18–20) and rat skeletal muscle (2). Moreover, mTORC1 can activate STAT3 and promote cell survival and growth in cancer cells (6). Taken together, these studies suggest a potential role for STAT3 in adaptive growth of skeletal muscle, though no studies have directly addressed its contribution to load-induced hypertrophy. To address this gap, we assessed the effects of 2 wk of synergist ablation (SA) on skeletal muscle hypertrophy in mice with skeletal muscle-specific STAT3 knockout (mKO) (21) and their floxed/wild-type (WT) littermates. In addition, we sought to assess the effects of different types of resistance exercise on STAT3 phosphorylation in human skeletal muscle.1
MATERIALS AND METHODS
Human study.
Skeletal muscle samples were obtained from the study recently published by McKendry et al. (9). Briefly, healthy male volunteers performed a single session of leg press-and leg extension resistance exercise [8 sets at 75% of one-repetition maximum (1RM) to volitional failure] with either 1 min (n = 8) or 5 min (n = 8) of interset rest. Skeletal muscle biopsies used in the present study were obtained at rest (Pre), 0 h, and 4 h postexercise (Post). Ethical approval was obtained through the NHS Black Country Research Ethics Committee (13/WM/0455); the study conformed to the standards set forward by the Declaration of Helsinki (7th version).
Animals.
STAT3 mKO mice were generated as previously described (21). All experiments were performed in 11-wk-old male mice, with floxed and Cre negative mice being the WT control mice. Cre expression was controlled by the muscle creatine kinase promoter. Breeding was designed such that all mice were floxed and were either Cre positive or negative (i.e., mice were littermates). Mice were housed under standard conditions with free access to food and water. All experiments were approved by and conducted in accordance with the Animal Care Program at the University of California, San Diego.
Synergist ablation.
Synergist ablation (SA) surgery, which induces overload of the plantaris, was performed as previously described (8). Briefly, mice were anesthetized with 2.5% isoflurane under sterile conditions, and the soleus and gastrocnemius were surgically removed. The contralateral nonoperated leg was used as control (CON). Two weeks after SA, muscles were collected from anesthetized animals.
Immunoblotting.
Plantaris homogenization and immunoblotting were performed as previously described (13), and proteins were detected with a primary antibody to 4E-BP1 (no. 9452, Cell Signaling), p-4E-BP1T37/46 (no. 9459, Cell Signaling), Akt (no. 4691, Cell Signaling), p-AktT308 (no. 2965, Cell Signaling), p-AktS473 (no. 4060, Cell Signaling), p-S6S235/S236 (no. 4858, Cell Signaling), p-S6S240/S244 (no. 5364, Cell Signaling), S6 (no. 2217, Cell Signaling), p-STAT3Y705 (no. 9145, Cell Signaling), STAT3 (no. 9132, Cell Signaling), and myostatin (ab71808, Abcam). Visualization and quantification were performed with G:BOX Chemi XT4 imager using GeneSys and GeneTools software (Syngene UK, Cambridge, UK).
Determination of muscle fiber cross-sectional area.
Muscle cross sections (10 μm) were taken from the muscle midbelly. Sections were first treated with 1% bovine serum albumin and normal goat and mouse serum as blocking agents. Sections were incubated overnight with a polyclonal anti-laminin antibody (dilution 1:1,000; Sigma, St. Louis, MO), and then with a secondary antibody, Alexa Fluor 594 goat anti-rabbit immunoglobulin G (dilution 1:200, Life Technologies, Carlsbad, CA). Slides were mounted with a hard-set mounting medium with DAPI (Vector, Burlingame, CA). Laminin was used to define the fiber perimeter and facilitate fiber area quantification. Sections were imaged with a Leica DM6000 microscope (Leica Microsystems, Buffalo Grove, IL) equipped with a Leica DFC365 FX camera (Leica Microsystems) using a ×10 objective and DAPI and TX2 filter sets. Fiber cross-sectional area (CSA) analysis and calculation were performed in a blinded manner with an automatic custom- written macro in ImageJ (National Institutes of Health, Bethesda, MD). To eliminate neurovascular structures and “optically fused” fibers, only regions with areas between 50 μm2 and 14,000 μm2 with circularity values greater than 0.3 were measured.
Statistical analysis.
Values are expressed as means ± SE (n = 6 muscles per group). Statistical significance was determined via two-way ANOVA with repeated measures and Bonferroni post hoc test. Significance was considered with a P value of <0.05.
RESULTS AND DISCUSSION
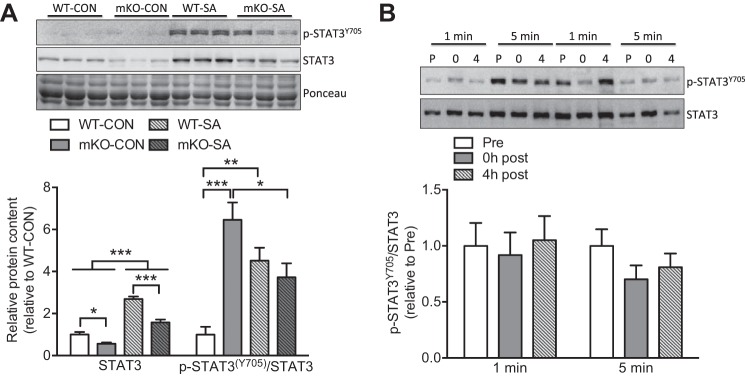
To date, the potential role of STAT3 in skeletal muscle adaptive growth is rather descriptive, implying an association between STAT3 activation and skeletal muscle hypertrophy (5, 18–20). To specifically define the contribution of STAT3 to adaptive skeletal muscle growth, we subjected WT and STAT3 mKO mice to a well-established model of skeletal muscle hypertrophy, the SA model. Chronic overload of plantaris significantly increased (4.5-fold) p-STAT3Y705 in WT mice (Fig. 1A). Interestingly, SA also augmented total STAT3 protein abundance in both WT and mKO mice (Fig. 1A). As we (21) and others (16, 24) have previously observed, while STAT3 was ~50% lower in mKO mice, STAT3 was still detected and, consequently, led to a misleading quantification of higher relative p-STAT3Y705 in mKO-CON mice (Fig. 1A). However, our recent paper demonstrated that this “residual” STAT3 protein is from nonmuscle cells, as immunoblotting of isolated muscle fibers revealed essentially complete knockout of STAT3 (21). Thus, we are confident that our model is appropriate for assessing the contribution of myofibril STAT3 to adaptive growth. In fact, SA reduced the relative levels of p-STAT3Y705 in mKO-SA as opposed to the increase observed in WT-SA muscles (Fig. 1A). Both skeletal muscle hypertrophy and regeneration strongly activate STAT3 in satellite cells (15, 17), suggesting that the nonmuscle p-STAT3Y705 detected in mKO potentially reflects the effect of SA on this cell population. Notably, it is possible that satellite cells are contributing nonrecombined DNA (and therefore, STAT3) to the adult myofiber, which could contribute to muscle hypertrophy. While we cannot discount this, we previously demonstrated that STAT3 is not present in isolated myofibers of mature muscle from the mKO mouse (21).
Fig. 1.
Synergist ablation (SA) increases signal transducer and activator of transcription 3 (STAT3) phosphorylation in mouse skeletal muscle. A: relative phosphorylation and total STAT3 protein content in mouse skeletal muscle following 2 wk of SA (n = 6 muscles per group). WT, wild type; mKO, skeletal muscle-specific STAT3 knockout. B: relative STAT3 phosphorylation levels in human skeletal muscle pre- (P), 0 h post- (0), and 4 h post- (4) resistance exercise with either 1 or 5 min of interset rest (n = 7–8 per group). All samples were derived at the same time and processed in parallel. Values are means ± SE. *P < 0.05, **P < 0.01, and ***P < 0.001.
To confirm that resistance exercise activates STAT3 in human skeletal muscle (18), we compared two resistance exercise protocols known to induce myofibrillar protein synthesis and mTORC1 signaling (9). Despite the anabolic response to resistance exercise with both 5 and 1 min of interset rest (9), neither protocol changed p-STAT3Y705 in human skeletal muscle (Fig. 1B). While this contrasts with previous work by Trenerry et al. (18), this difference is likely due to when muscle was sampled after exercise; we sampled muscle immediately and 4 h postexercise, whereas Trenerry et al. found that pSTAT3 peaked 2 h after exercise but returned to baseline by 4 h. Our data do not rule out the activation of STAT3 in human skeletal muscle following resistance exercise; the data further suggest that STAT3 would be transiently activated, as reported by Trenerry et al. (18).
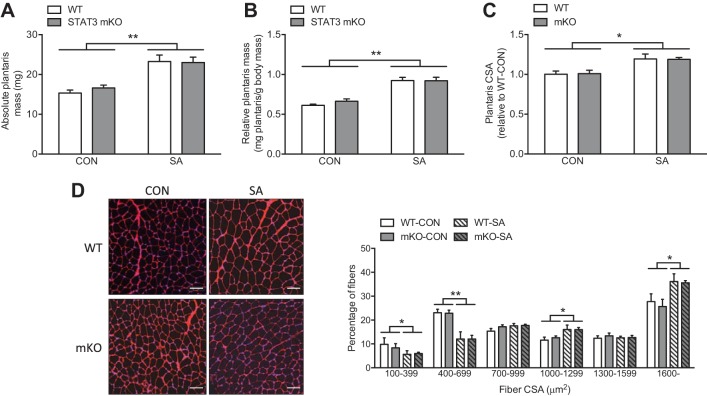
Under control conditions, WT and mKO mice had comparable plantaris mass (Fig. 2, A and B), which is consistent with the lack of effect of STAT3 mKO or inhibition on basal body composition and skeletal muscle mass (21, 24). Interestingly, despite the positive role of STAT3 in skeletal muscle wasting (16, 24), the robust increase in both absolute and relative plantaris mass induced by SA was not impaired in STAT3 mKO mice (Fig. 2, A and B). To specifically assess hypertrophy, we measured average plantaris fiber size, which revealed a comparable increase in muscle fiber CSA in WT-SA and mKO-SA (Fig. 2C). Analysis of fiber size distribution reflected a consistent switch toward a higher proportion of muscle fibers with large CSA and a decrease in smaller muscle fibers (Fig. 2D). However, in line with muscle mass and average CSA data, the effects of SA on fiber size distribution were not different between genotypes (Fig. 2D). Interestingly, Bonetto et al. (3) found that transient STAT3 loss of function promoted hypertrophy both in cultured myotubes and in mouse skeletal muscle, which the authors speculated was dependent on interleukin-6 (IL-6) inhibition. Contrasting this, however, IL-6 knockout mice exhibit normal skeletal muscle fiber CSA under basal conditions and, though not fully understood, adaptive growth appears to be abnormal compared with wild-type mice (15, 22). Therefore, although the effects of transient STAT3 inactivation on adaptive growth and anabolic signaling remain to be studied, our data demonstrate that mice with chronic loss of STAT3 have no impairments in skeletal muscle adaptations to load-induced growth.
Fig. 2.
Synergist ablation (SA) increases skeletal muscle mass and fiber size. A: absolute mass of both control (CON) and SA plantaris. B: relative mass of both CON and SA plantaris. C: changes in average plantaris fiber cross-sectional area (CSA) following SA. D: representative images (red: laminin, blue: nuclei) and quantification of skeletal muscle fiber size (CSA) distribution of both CON and SA plantaris (white scale bar = 100 μm). Values are means ± SE (n = 6 muscles per group). *P < 0.05, **P < 0.01.
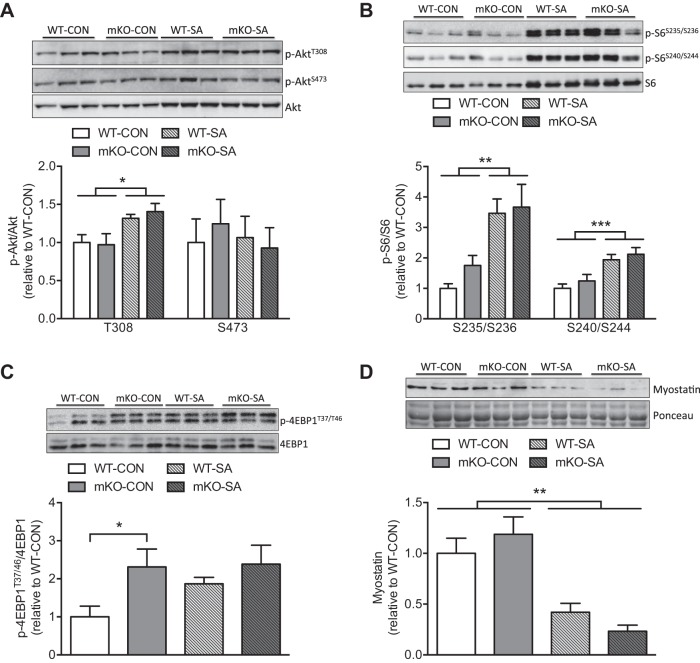
To understand whether canonical growth signaling in response to SA was altered by loss of STAT3, we assessed different PI3K and mTORC1 target proteins. SA increased PI3K signaling in both genotypes, as reflected by higher relative phosphorylation levels of AktT308, whereas no effect on mTORC2-mediated AktS473 phosphorylation was observed (Fig. 3A). Furthermore, activation of the mTORC1 pathway was clearly reflected by higher levels of p-S6S235/S236 and p-S6S240/S44, with comparable responses in WT-SA and mKO-SA mice (Fig. 3B). 4E-BP1 (a direct target of mTORC1) phosphorylation was higher in mKO-CON and trended to increase only in WT-SA muscles (Fig. 3C). The higher p-4E-BP1T37/46 and trend to higher p-S6S235/S236 in mKO-CON mice suggest that STAT3 might exert a negative effect on mTORC1 activity. Besides the positive effects of mTORC1 on STAT3 transcriptional activity (6), a possible negative feedback loop has not been fully elucidated. A mechanism for such negative feedback potentially involves STAT3-mediated upregulation of the tumor suppressor phosphatase and tensin homolog, which subsequently inhibits Akt activation (23). Moreover, IL-6-mediated activation of STAT3 is associated with inhibition of mTORC1 signaling (11). However, we did not find changes in Akt phosphorylation; instead, our data suggest that the enhanced 4E-BP1 and S6 phosphorylation observed in mKO-CON does not translate into greater skeletal muscle mass or fiber size. The possible negative feedback mediated by STAT3 on mTORC1 signaling and its biological function requires further investigation.
Fig. 3.
Synergist ablation (SA) promotes the activation of anabolic pathways. A–C: relative phosphorylation levels of downstream targets of phosphoinositide 3-kinase (PI3K) (AktT308), mechanistic target of rapamycin complex 2 (mTORC2) (AktS473), and mTORC1-S6K1 (S6S235/S236 and S6S240/S244) and mTORC1 (4E-BP1T37/46) in both control (CON) and SA plantaris. D: total protein levels of the procatabolic protein myostatin under CON and SA conditions. All samples were derived at the same time and processed in parallel. Values are means ± SE (n = 6 muscles per group). *P < 0.05, **P < 0.01, and ***P < 0.001.
STAT3 has been shown to indirectly promote myostatin expression and catabolic signaling in the context of skeletal muscle wasting (24). As expected, the protein content of myostatin was strongly reduced by SA, though without additional effects of STAT3 mKO (Fig. 3D). Moreover, similar to studies by others (16, 24), STAT3 KO did not modify basal myostatin protein levels in skeletal muscle (Fig. 3D). While myostatin mRNA levels are downregulated by SA, which implies a transcriptional regulation (12), our data suggest that this is not mediated by STAT3.
Altogether, the present study demonstrates that under basal conditions, genetic deletion of STAT3 does not impact skeletal muscle mass, thus consistent with previous reports demonstrating that STAT3 does not regulate skeletal muscle mass during development or in the absence of pathological stimuli (16, 21, 24). Furthermore, in the context of skeletal muscle hypertrophy, we found that STAT3 is not required for SA-mediated activation of anabolic signaling pathways such as PI3K and mTORC1, nor is it required for skeletal muscle adaptive growth in healthy mice.
GRANTS
This publication was supported in part by a Sir Henry Wellcome Post-doctoral Fellowship (103094/Z/13/Z; to J. Pérez-Schindler), Biotechnology and Biological Sciences Research Council New Investigator Award (BB/L023547/1 to A. Philp), and National Institutes of Health Grant R01 AG043120 (to S. Schenk).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
J.P.-S., A.P., and S.S. conceived and designed research; J.P.-S., M.C.E., J.M., and L.B. performed experiments; J.P.-S. analyzed data; J.P.-S., A.P., and S.S. interpreted results of experiments; J.P.-S. prepared figures; J.P.-S. and S.S. drafted manuscript; J.P.-S., M.C.E., J.M., L.B., A.P., and S.S. approved final version of manuscript; M.C.E., J.M., L.B., and A.P. edited and revised manuscript.
ACKNOWLEDGMENTS
We thank Sarah Nalbandian and Chandra Inglis at UC San Diego for excellent technical assistance.
Present address: J. Pérez-Schindler, Biozentrum, University of Basel, Basel, Switzerland.
Footnotes
This article is the topic of an Editorial Focus by Adam J. Amorese and Espen E. Spangenburg (1).
REFERENCES
- 1.Amorese AJ, Spangenburg EE. Defining the STATus quo in muscle hypertrophy. Focus on “Overload-mediated skeletal muscle hypertrophy is not impaired by loss of myofiber STAT3.” Am J Physiol Cell Physiol. doi: 10.1152/ajpcell.00165.2017. [DOI] [PubMed] [Google Scholar]
- 2.Begue G, Douillard A, Galbes O, Rossano B, Vernus B, Candau R, Py G. Early activation of rat skeletal muscle IL-6/STAT1/STAT3 dependent gene expression in resistance exercise linked to hypertrophy. PLoS One 8: e57141, 2013. doi: 10.1371/journal.pone.0057141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonetto A, Aydogdu T, Jin X, Zhang Z, Zhan R, Puzis L, Koniaris LG, Zimmers TA. JAK/STAT3 pathway inhibition blocks skeletal muscle wasting downstream of IL-6 and in experimental cancer cachexia. Am J Physiol Endocrinol Metab 303: E410–E421, 2012. doi: 10.1152/ajpendo.00039.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen S, Nathan JA, Goldberg AL. Muscle wasting in disease: molecular mechanisms and promising therapies. Nat Rev Drug Discov 14: 58–74, 2015. doi: 10.1038/nrd4467. [DOI] [PubMed] [Google Scholar]
- 5.Drummond MJ, Fry CS, Glynn EL, Timmerman KL, Dickinson JM, Walker DK, Gundermann DM, Volpi E, Rasmussen BB. Skeletal muscle amino acid transporter expression is increased in young and older adults following resistance exercise. J Appl Physiol (1985) 111: 135–142, 2011. doi: 10.1152/japplphysiol.01408.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laplante M, Sabatini DM. Regulation of mTORC1 and its impact on gene expression at a glance. J Cell Sci 126: 1713–1719, 2013. doi: 10.1242/jcs.125773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marcotte GR, West DW, Baar K. The molecular basis for load-induced skeletal muscle hypertrophy. Calcif Tissue Int 96: 196–210, 2015. doi: 10.1007/s00223-014-9925-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McGee SL, Mustard KJ, Hardie DG, Baar K. Normal hypertrophy accompanied by phosphoryation and activation of AMP-activated protein kinase alpha1 following overload in LKB1 knockout mice. J Physiol 586: 1731–1741, 2008. doi: 10.1113/jphysiol.2007.143685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKendry J, Pérez-López A, McLeod M, Luo D, Dent JR, Smeuninx B, Yu J, Taylor AE, Philp A, Breen L. Short inter-set rest blunts resistance exercise-induced increases in myofibrillar protein synthesis and intracellular signalling in young males. Exp Physiol 101: 866–882, 2016. doi: 10.1113/EP085647. [DOI] [PubMed] [Google Scholar]
- 10.McLeod M, Breen L, Hamilton DL, Philp A. Live strong and prosper: the importance of skeletal muscle strength for healthy ageing. Biogerontology 17: 497–510, 2016. doi: 10.1007/s10522-015-9631-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pelosi M, De Rossi M, Barberi L, Musarò A. IL-6 impairs myogenic differentiation by downmodulation of p90RSK/eEF2 and mTOR/p70S6K axes, without affecting AKT activity. BioMed Res Int 2014: 206026, 2014. doi: 10.1155/2014/206026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pérez-Schindler J, Summermatter S, Santos G, Zorzato F, Handschin C. The transcriptional coactivator PGC-1α is dispensable for chronic overload-induced skeletal muscle hypertrophy and metabolic remodeling. Proc Natl Acad Sci USA 110: 20314–20319, 2013. doi: 10.1073/pnas.1312039110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Philp A, Chen A, Lan D, Meyer GA, Murphy AN, Knapp AE, Olfert IM, McCurdy CE, Marcotte GR, Hogan MC, Baar K, Schenk S. Sirtuin 1 (SIRT1) deacetylase activity is not required for mitochondrial biogenesis or peroxisome proliferator-activated receptor-gamma coactivator-1alpha (PGC-1alpha) deacetylation following endurance exercise. J Biol Chem 286: 30561–30570, 2011. doi: 10.1074/jbc.M111.261685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Powers SK, Lynch GS, Murphy KT, Reid MB, Zijdewind I. Disease-Induced Skeletal Muscle Atrophy and Fatigue. Med Sci Sports Exerc 48: 2307–2319, 2016. doi: 10.1249/MSS.0000000000000975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Serrano AL, Baeza-Raja B, Perdiguero E, Jardí M, Muñoz-Cánoves P. Interleukin-6 is an essential regulator of satellite cell-mediated skeletal muscle hypertrophy. Cell Metab 7: 33–44, 2008. doi: 10.1016/j.cmet.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 16.Silva KA, Dong J, Dong Y, Dong Y, Schor N, Tweardy DJ, Zhang L, Mitch WE. Inhibition of Stat3 activation suppresses caspase-3 and the ubiquitin-proteasome system, leading to preservation of muscle mass in cancer cachexia. J Biol Chem 290: 11177–11187, 2015. doi: 10.1074/jbc.M115.641514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tierney MT, Aydogdu T, Sala D, Malecova B, Gatto S, Puri PL, Latella L, Sacco A. STAT3 signaling controls satellite cell expansion and skeletal muscle repair. Nat Med 20: 1182–1186, 2014. doi: 10.1038/nm.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trenerry MK, Carey KA, Ward AC, Cameron-Smith D. STAT3 signaling is activated in human skeletal muscle following acute resistance exercise. J Appl Physiol (1985) 102: 1483–1489, 2007. doi: 10.1152/japplphysiol.01147.2006. [DOI] [PubMed] [Google Scholar]
- 19.Trenerry MK, Carey KA, Ward AC, Farnfield MM, Cameron-Smith D. Exercise-induced activation of STAT3 signaling is increased with age. Rejuvenation Res 11: 717–724, 2008. doi: 10.1089/rej.2007.0643. [DOI] [PubMed] [Google Scholar]
- 20.Trenerry MK, Della Gatta PA, Larsen AE, Garnham AP, Cameron-Smith D. Impact of resistance exercise training on interleukin-6 and JAK/STAT in young men. Muscle Nerve 43: 385–392, 2011. doi: 10.1002/mus.21875. [DOI] [PubMed] [Google Scholar]
- 21.White AT, LaBarge SA, McCurdy CE, Schenk S. Knockout of STAT3 in skeletal muscle does not prevent high-fat diet-induced insulin resistance. Mol Metab 4: 569–575, 2015. doi: 10.1016/j.molmet.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White JP, Reecy JM, Washington TA, Sato S, Le ME, Davis JM, Wilson LB, Carson JA. Overload-induced skeletal muscle extracellular matrix remodelling and myofibre growth in mice lacking IL-6. Acta Physiol (Oxf) 197: 321–332, 2009. doi: 10.1111/j.1748-1716.2009.02029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zha X, Hu Z, He S, Wang F, Shen H, Zhang H. TSC1/TSC2 inactivation inhibits AKT through mTORC1-dependent up-regulation of STAT3-PTEN cascade. Cancer Lett 313: 211–217, 2011. doi: 10.1016/j.canlet.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Zhang L, Pan J, Dong Y, Tweardy DJ, Dong Y, Garibotto G, Mitch WE. Stat3 activation links a C/EBPδ to myostatin pathway to stimulate loss of muscle mass. Cell Metab 18: 368–379, 2013. doi: 10.1016/j.cmet.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]