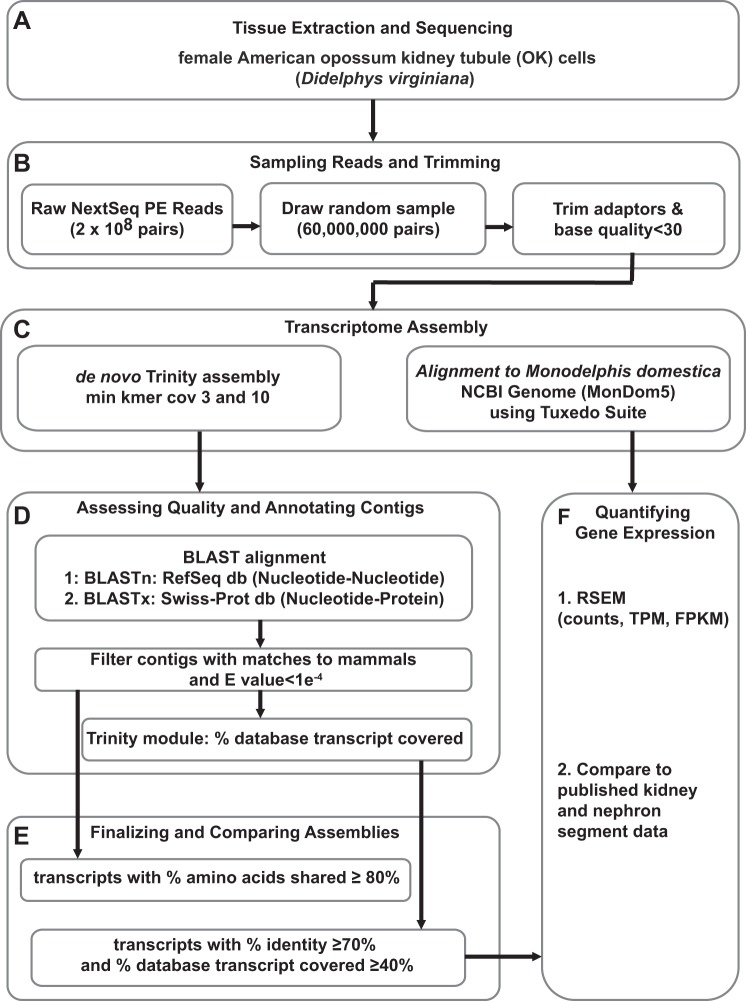
Fig. 3.
Pipeline for transcriptome assembly and analysis. A: single tissue samples were sequenced using NextGen sequencing technology to generate >200 million read pairs. B: 60 million randomly selected pairs were extracted and trimmed for adaptors and bases with quality score <30. C: reads were assembled using 2 methods: Trinity de novo assembly and alignment to the Monodelphis domestica NCBI reference genome (GCF_000002295.2 MonDom5) using TopHat and Cufflinks in the Tuxedo Suite. We tested parameters to determine which provides the most complete de novo assembly. D: de novo assembled contigs were aligned to the nucleotide RefSeq RNA database and to the protein Swiss-Prot database using BLASTn and BLASTx, respectively. The BLAST output against these databases was used to filter contigs that matched to mammals with an e-value <1e−4 and to annotate contigs with gene names. Using the Trinity provided module, we calculated the percent coverage of the top database match for each contig. E: from all assembled contigs, we utilized the various statistics to filter our assemblies, one whose amino acids matches identically to greater than or equal to 80% of their top database hit, and a second less stringent filter of contigs with at %Identity >70% and percent_hit_length >40% to include high-quality but partial transcript assemblies. F: the contigs in the filtered de novo assembly and the alignment to MonDom5 were quantified at the isoform and gene level using RSEM to generate counts, transcripts per million (TPM), and fragments per kilobase per million (FPKM). With the use of FPKM, the expression patterns were compared with published studies of rat kidney tubule expression.