Abstract
Acylglycerophosphate acyltransferase 4 (AGPAT4)/lysophosphatidic acid acyltransferase delta catalyzes the formation of phosphatidic acid (PA), a precursor of triacylglycerol (TAG). We investigated the effect of Agpat4 gene ablation on white adipose tissue (WAT) after finding consistent expression across depots. Epididymal WAT mass was 40% larger in male Agpat4−/− mice than wild-type littermates, but unchanged in perirenal, retroperitoneal, and inguinal WAT and subscapular brown adipose tissue. Metabolic changes were identified in epididymal WAT that were not evident in perirenal WAT, which was analyzed for comparison. The total epididymal TAG content doubled, increasing adipocyte cell size without changing markers of differentiation. Enzymes involved in de novo lipogenesis and complex lipid synthesis downstream of phosphatidic acid production were also unchanged. However, total epididymal TAG hydrolase activity was reduced, and there were significant decreases in total ATGL and reduced phosphorylation of hormone-sensitive lipase at the S563 and S660 PKA-activation sites. Analysis of Agpats 1, 2, 3, and 5, as well as Gpats 1, 2, 3, and 4, demonstrated compensatory upregulation in perirenal WAT that did not occur in epididymal WAT. Our findings therefore indicate depot-specific differences in the redundancy of Agpat4 and highlight the molecular and metabolic heterogeneity of individual visceral depots.
Keywords: acylglycerophosphate acyltransferase 4, adipose tissue, lipid biochemistry, lipolysis and fatty acid metabolism, phospholipids/phosphatidic acid, triglycerides, lipogenesis, lysophosphatidic acid acyltransferase
The acylglycerophosphate acyltransferase/lysophosphatidic acid acyltransferase (AGPAT/LPAAT) family is a group of proteins that have been identified based on sequence homology (1, 2). Each member contains two highly conserved catalytic motifs, NHX4D and a downstream proline residue, as well as two substrate binding motifs, EGTR and FX2R. Currently, 11 AGPAT family members have been identified. True AGPAT/LPAATs function in the second committed step of the Kennedy pathway for the de novo biosynthesis of glycerophospholipids and triacylglycerol (TAG) by catalyzing the acyl-CoA-dependent formation of phosphatidic acid (PA) using lysophosphatidic acid (LPA) as the preferred acyl acceptor (3). Functional characterization of these enzymes has, however, narrowed the group of true AGPATs/LPAATs that prefer LPA to only 5 of the 11 isoforms (AGPATs 1, 2, 3, 4, and 5), with the rest primarily utilizing a different lysophospholipid acyl-acceptor, or glycerol-3-phosphate (2).
These AGPAT/LPAAT isoforms show different tissue distribution patterns, with AGPATs 1 and 3 demonstrating ubiquitious expression and AGPATs 2, 4, and 5 showing distinct tissue-specific profiles and regulation by fasting (4, 5). The physiological significance of these distributions, the role of each isoform in different tissues, and the reason for the apparently redundant expression of homologous enzymes in the same tissue is not well understood (6). Recently, our laboratory sought to functionally characterize AGPAT4 in a series of in vitro studies, and to understand the in vivo biochemical role of AGPAT4 in the brains of mice (7). Although it was determined that AGPAT4 functions catalytically as a true AGPAT/LPAAT in vitro, utilizing only LPA as its major lyosphospholipid acyl-acceptor, it was also found that AGPAT4 specifically supports the production of brain phosphatidylinositol (PI), phosphatidylcholine (PC), and phosphatidylethanolamine (PE) (7). This suggested that in mouse brains, AGPAT4-derived PA forms a functionally distinct substrate pool for the synthesis of these specific downstream phospholipid species (7). As a result, these findings aid in explaining both the distinct tissue expression patterns and apparent redundancy of Agpat/Lpaat isoform expression in vivo. It remains to be determined, however, whether AGPAT4 functions to support PC, PE, and PI levels in other tissues, as it does in brain, or whether it has diverse, tissue-specific roles.
Agpat4 is relatively abundant in murine renal white adipose tissue (WAT) (4). Several AGPAT isoforms are known to be expressed in WAT, and have been linked to the regulation of TAG production, the differentiation of adipocytes, and the modulation of various pathologies including lipodystrophy, diabetes, and hepatic steatosis (8–12). In particular, Agpat2 and Agpat6 are highly expressed in WAT, and the role of these isoforms has been studied in this tissue using overexpression and gene ablation models (8–12). AGPAT6 has been show to play a criticial physiological role in the accumulation of TAG in WAT, as Agpat6−/− mice have reduced adipocyte TAG content, smaller epididymal adipocyte size, and subcutaneous lipodystrophy (12). The role of AGPAT2 in the production and regulation of TAG synthesis was discovered originally from studies in patients with AGPAT2 mutations who have a form of congenital generalized lipodystrophy known as Berardinelli-Seip syndrome. This condition is characterized by a lack of adipose tissue, insulin resistance, hypertriglyceridemia, hepatic steatosis, and early onset diabetes (8). Although the exact mechanism by which AGPAT2 mutations cause lipodystrophy has yet to be elucidated, AGPAT2 has been shown to be imperative for both lipogenesis and adipogenesis. Agpat2 is upregulated 40-fold during adipocyte differentiation, and Agpat2 knockdown results in delayed adipocyte differentiation and impaired TAG synthesis and storage (11).
Current understanding of the role of AGPATs in WAT biology is incomplete, with no specific studies focused on the function of AGPATs 1, 3, 4, or 5 in WAT. We therefore used an Agpat4 mouse gene ablation model (Agpat4−/−) to investigate the role of AGPAT4 in WAT function. In our initial investigations, we found that the epididymal adipose depot mass was larger in male Agpat4−/− mice compared with wild-type littermates, but that the mass of perirenal, retroperitoneal, and inguinal WAT, as well as subscapular brown adipose tissue (BAT), was not different. We therefore investigated lipogenesis, lipolysis, and adipocyte differentiation in epididymal WAT and, for comparison, perirenal WAT. Measures of whole body metabolism were also performed.
MATERIALS AND METHODS
Animals
All animal procedures were approved by the University of Waterloo Animal Care Committee. Mice were housed in a temperature and humidity controlled environment on a 12:12 h light/dark cycle. Standard rodent chow and water were provided ad libitum. Heterozygous Agpat4+/− mice were cyrorevived at the Mutant Mouse Regional Resource Center from a strain produced by Lexicon Genetics/Genentech (B6;129S5-Agpat4tm1Lex/Mmucd). Exons 4, 5, and 6 of Agpat4 (NCBI accession NM_026644.1) were replaced in mouse 129S5 embryonic stem cells with a LacZ/Neo cassette, and we have previously reported confirmation of genotype (7). Progeny from two clones (1A9 and 1F9) achieved germline transmission after crossing with C57BL/6J mice. Cryorevived heterozygous offspring were sent from the University of California at Davis to the University of Waterloo. Male and female heterozygous littermates were intercrossed to achieve wild-type, heterozygous, and homozygous progeny. Adult male and female Agpat4−/− mice, aged 9–12 weeks old, were used with age-matched wild-type littermates as controls. DNA was isolated from ear punches, and genotyping was determined by visualization of amplicons of the correct size using primers specific for a region present only in the wild-type animals (forward: 5′-TTA GCA TAG TGG GCG AAG TTC-3′, reverse: 5′-GGT AGT GGC CAA GTT AAT AGT CCT-3′ 216 bp) and primers specific to the recombined targeted region (forward: 5′-GCA GCG CAT CGC CTT CTA TC-3′, reverse: 5′-CTC CCA TTT CTA GGA AGG AAG CAG-3′ 344 bp) as previously described (7). RT-PCR showed the expected absence of Agpat4 trancripts in mouse WAT depots (Fig. 1B) using a forward primer in the excised fourth exon (5′-ATC ACG CTG ACT GCT ACG TTC GGA-3′) and a reverse primer in the excised sixth exon (5′-GAG TCT TCT GGG AAG ACC CCT GTC-3′). Amplification was performed using 1 μl cDNA in a T100 Thermal Cycler (Bio-Rad, Hercules, CA) with the following conditions: 94°C for 5 min, 39 cycles of 94°C for 1 min, 58°C for 1 min, 72°C for 30 s, followed by 72°C for 10 min.
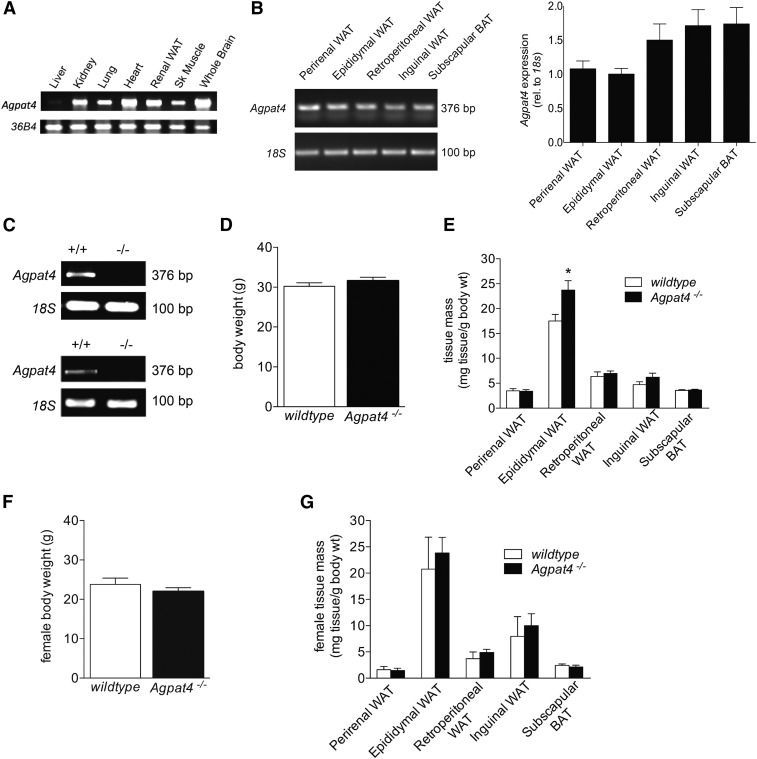
Fig. 1.
Agpat4 is expressed in multiple adipose tissue depots, but only epididymal WAT mass is affected by gene knockout. A: Agpat4 mRNA expression determined by RT-PCR in different tissues from male C57Bl/6J mice (n = 3). B: Agpat4 mRNA expression determined by RT-PCR in multiple adipose tissue depots from male C57Bl/6J mice. Left, Representative ethidium bromide gel and right, quantification of expression (n = 3). C: RT-PCR showing absence of Agpat4 transcript in epididymal (top) and perirenal WAT (bottom) from knockout mice. D: Body weights of male wild-type and littermate Agpat4−/− mice at 9–11 weeks of age (n = 25–28). E: Adipose depot weights from male mice normalized to body mass (n = 20–34). F: Body weights of female wild-type and littermate Agpat4−/− mice at 9–11 weeks of age (n = 5). G: Weights of adipose tissues from female mice normalized to body mass (n = 5). Data are means ± SEM; *P < 0.05 versus wild-type.
Indirect calorimetry
To obtain measurements of respiratory exchange ratio (RER), heat production, daily average oxygen consumption (VO2), food intake, fat oxidation, carbohydrate oxidation, total activity, dual beam movement, and Z-axis movement, an Oxymax Comprehensive Laboratory Animal Monitoring System (Columbus Instruments, Columbus, OH) was used as previously described (13) for two trials of 72 h each. Animals were housed individually, and a 2 h acclimation period in the metabolic chamber was performed prior to monitoring over a 72 h period under consistent environmental temperature (22°C). RER was calculated using the ratio of carbon dioxide to VO2. Heat was determined using the product of the calorific value (derived from the RER) and volume of oxygen consumed. Food consumption was determined by monitoring chow weight, and activity was determined through infra-red beam breaks.
RNA extraction, RT-PCR, and qPCR
Total RNA was isolated using TRIzol® reagent according to the manufacturer’s instructions (Sigma-Aldrich, Oakville, ON), and 2.5 μg was reverse transcribed to cDNA using a High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, Waltham, MA) in a Bio-Rad T100 thermal cycler using the following settings: 25°C for 10 min, 37°C for 2 h, and 85°C for 5 s. Semi-quantitative PCR was performed in a Bio-Rad T100 thermal cycler using 1 μl cDNA (amplification primers are listed in Table 1) and the following cycling conditions: one cycle of 94°C for 4 min followed by 30 cycles of 94°C for 30 s, 58°C for 30 s, 72°C for 1 min, followed by a final extension of 72°C for 10 min. Amplicons were resolved in a 1% agarose Tris-acetate-EDTA gel with ethidium bromide for visualization under UV light. Quantitative real time (q) PCR was performed using a Bio-Rad CFX96 Touch™ Real-Time PCR Detection System with SsoFast EvaGreen supermix (Bio-Rad). Amplification was performed under the following conditions: 95°C for 2 min, 40 cycles of 95°C for 10 s, 60°C for 20 s, followed by a melt curve to verify a single amplicon. Relative quantification was assessed using the ΔΔCt method. Sequences for qPCR primers are also listed in Table 1.
TABLE 1.
PCR primers
| Gene | Direction | Sequence | Product Size (bp) |
| Agpat1 | Forward | 5′-AGA CCT TGC TCA CCC AGG AT-3′ | 134 |
| Reverse | 5′-GAT GGG GAT GAT GGG GAC CT-3′ | ||
| Agpat2 | Forward | 5′-CCGTGGTGTACTCGTCTTTCT-3′ | 107 |
| Reverse | 5′-CAGACCATTGGTAGGGACAGC-3′ | ||
| Agpat3 | Forward | 5′-GCT TCG TCC TGG GTG TCT TT-3′ | 137 |
| Reverse | 5′-GTT GCC ATA GCT GGA GCC TT-3′ | ||
| Agpat5 | Forward | 5′-GGA CAT GTG CGC TAC GTA CT-3′ | 167 |
| Reverse | 5′-AGA TAC ATC GGT GTT CCT GCG-3′ | ||
| Cdipt | Forward | 5′-GATCGACCTGTCTGGGAACC-3′ | 78 |
| Reverse | 5′-TTTCCAGCACACAGGGTGAA-3′ | ||
| Cds1 | Forward | 5′-TGGACATGGCGGGATAATGG-3′ | 111 |
| Reverse | 5′-TGGAGCACTTTGCTGGGATT-3′ | ||
| Cds2 | Forward | 5′-GGGTTCTTCGCCAGTGGATT-3′ | 89 |
| Reverse | 5′-AAGCGATCCATGATGCCTCC-3′ | ||
| C/ebpα | Forward | 5′-GCAAAGCCAAGAAGTCGGTG-3′ | 114 |
| Reverse | 5′-TCTCCACGTTGCGTTGTTTG-3′ | ||
| C/ebpβ | Forward | 5′-AAGCTGAGCGACGAGTACAAGA-3′ | 116 |
| Reverse | 5′-GTCAGCTCCAGCACCTTGTG-3′ | ||
| Cept1 | Forward | 5′-TTGTACTGTGGCAGGGACCA-3′ | 97 |
| Reverse | 5′-TGTTCCTGCTATGGTTGACCC-3′ | ||
| Chpt1 | Forward | 5′-ACTGTCTTTATTGGGCCAGGT-3′ | 90 |
| Reverse | 5′-GACCATTGCTATCCACAGAACA-3′ | ||
| Dgat1 | Forward | 5′-CTGGATTGTGGGCCGATTCT-3′ | 94 |
| Reverse | 5′-ATACATGAGCACAGCCACCG-3′ | ||
| Dgat2 | Forward | 5′-AAGACATCGACCTGTACCATGC-3′ | 92 |
| Reverse | 5′-CTCAGTCTCTGGAAGGCCAAA-3′ | ||
| Fabp4 | Forward | 5′-GTGGGATGGAAAGTCGACCA-3′ | 70 |
| Reverse | 5′-CATAACACATTCCACCACCAGC-3′ | ||
| Fat/cd36 | Forward | 5′-ACTGTGGCTAAATGAGACTGGG-3′ | 93 |
| Reverse | 5′-ACCATGCCAAGGAGCTTGAT-3′ | ||
| Gpat1 | Forward | 5′-CTGCAGACGCCGCTGG-3′ | 179 |
| Reverse | 5′-AACCCACAGTCAACCCAGTC-3′ | ||
| Gpat2 | Forward | 5′-ACACTTGTTGCAGCCCATTG-3′ | 161 |
| Reverse | 5′-CCACACTGGCACTCAGAACA-3′ | ||
| Gpat3 | Forward | 5′-CACGGTGGATTGATGGGGAT-3′ | 173 |
| Reverse | 5′-TTGATGCAAGTACCTTCTGGGA-3′ | ||
| Gpat4 | Forward | 5′-GGTGGCTAAGAGGCTGACTG-3′ | 155 |
| Reverse | 5′-ACTTGATAGCCACAGGGTAAACA-3′ | ||
| Il-1β | Forward | 5′-cctgctggtgtgtgacgttccc-3′ | 84 |
| Reverse | 5′-gggtccgacagcacgaggct-3′ | ||
| Il-6 | Forward | 5′-gctggagtcacagaaggagtggct-3′ | 79 |
| Reverse | 5′-ggcataacgcactaggtttgccgag-3′ | ||
| Iplaβ | Forward | 5′-CCTTTCCATTACGCTGTGCAA-3′ | 103 |
| Reverse | 5′-GACTCACGGCTTGGTTGTT-3′ | ||
| Iplaγ | Forward | 5′-CAAAGACAAGAAGGCAGAGGAG-3′ | 106 |
| Reverse | 5′-TAAGGCCTGAACTAAGGCTCG-3′ | ||
| Lpin1 | Forward | 5′-CCTTAGGGAGCCGGAAGACT-3′ | 91 |
| Reverse | 5′-ATTGTTGGCGACTGGTCACT-3′ | ||
| Pgs1 | Forward | 5′-GGGTCCAGCTCCAGGAATAC-3′ | 111 |
| Reverse | 5′-TAGGAGAGCCAATCAGCGTG-3′ | ||
| Pparγ | Forward | 5′-CACAATGCCATCAGGTTTGG-3′ | 82 |
| Reverse | 5′-GCTGGTCGATATCACTGGAGATC-3′ | ||
| Pref1 | Forward | 5′-GACAGGCCATCTGCTTCACC-3′ | 116 |
| Reverse | 5′-GTTGTAGCGCAGGTTGGACA-3′ | ||
| TNFα | Forward | 5′-caacgccctcctggccaacg-3′ | 114 |
| Reverse | 5′-tcggggcagccttgtccctt-3′ | ||
| 18s | Forward | 5′-GATCCATTGGAGGGCAAGTCT-3′ | 79 |
| Reverse | 5′-AACTGCAGCAACTTTAATATACGCTATT-3′ |
Adipocyte size determination
Epididymal and perirenal WAT samples were collected from Agpat4−/− and wild-type control littermates and embedded in paraffin for sectioning and staining, as previously described (14, 15). Briefly, fresh tissues were fixed in 4% paraformaldehyde at 4°C for 48 h, dehydrated, and embedded in paraffin for microtome sectioning into 6 μm thick slices that were carefully transferred to microscope slides, and allowed to adhere at 40°C overnight. Sections were then rehydrated and stained with hematoxylin and eosin as previously described (14, 15). Five separate fields from four different mice were quanitified using Image J software to derive an estimate of adipocyte size (16).
Immunoblotting
WAT depots were homogenized in lysis buffer (50 mM Tris, pH 7.4, 1 mM EDTA, 0.1 M sucrose, 5 mM sodium fluoride, 10 mM sodium orthovanadate, with 1% protease inhibitor cocktail) and mixed with 6 × protein loading dye (10 mM Tris HCl, 9% SDS, 50% glycerol, 0.03% bromophenol blue) followed by denaturation for 5 min at 95°C. Samples were electrophoresed through 10% TGX Stain-Free FastCast Acrylamide gels (Bio-Rad, Mississauga, Ontario, Canada) and blocked with 5% blocker (w/v) in TBST (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.1% Tween-20) for 1 h followed by incubation overnight at 4°C with primary antibody (1:1,000) in 1% blocker (w/v) (all antibodies from Cell Signaling, Beverly, MA). Membranes were imaged by chemiluminescence using LuminataTM Crescendo HRP substrate in a Bio-Rad ChemiDocTM Touch Imaging system. Equal protein loading was determined by imaging and quantification of total protein content per lane visualized in TGX Stain-Free gels at 340 nM.
Lipid extraction, TLC, and GC
Total lipids were extracted from epididymal and perirenal WAT depots from wild-type and Agpat4−/− mice as described by Folch, Lees, and Sloane Stanley (17). The organic layer was collected and the sample was reextracted with 2 ml chloroform. The samples were centrifuged at 1,734 g for 5 min and the organic layer was collected and pooled with the previous collection. Lipid extracts were dried down and applied to a silica gel G plate (Analtech, Newark, De) and resolved in a TLC tank to separate neutral lipids with heptane:diethyl ether:glacial acetic acid (80:20:2, v/v/v). The band corresponding to TAG, as determined by comparison to a known lipid standard (Nu Chek Prep, Elysian, MN), was scraped for analysis. The band corresponding to polar lipids was also scraped, and lipids were extracted twice from the silica using 2:1 chloroform:methanol (v/v) and the extract was applied to a silica gel H plate (Analtech) for further resolution of individual phospholipid classes using a solvent mixture of chloroform:methanol:isopropanol:0.25% KCl:triethylamine (30:9:25:6:18, v/v/v/v/v). The bands corresponding to these phospholipid classes were identified using known standards (Avanti Polar Lipids, Alabaster, AL) and scraped. Analysis of FA content and composition of complex lipids was performed by GC with flame ionization detection as previously described (18). Briefly, lipids were extracted from the silica with 2:1 chloroform:methanol that included an internal standard (10 μg 22:3n-3 methyl ester per ml) and an antioxidant (butylated hydroxytoluene, Thermo Fisher Scientific) and extracts were dried down under a stream of N2 gas. Fatty acyl chains on lipids were transesterified to FA methyl esters using 14% boron trifluoride in methanol (Sigma-Aldrich) with hexane on a 95°C heat block for 1 h. Analysis was then performed using a Varian 3900 gas chromatograph equipped with a DB-FFAP 15 m × 0.1 mm injected dose × 0.10 μm film thickness, nitroterephthalic acid modified, polyethylene glycol, capillary column (J&W Scientific from Agilent Technologies, Mississauga, ON) with hydrogen as the gas carrier. A volume of 1 μl of each sample was introduced by a Varian CP-8400 autosampler into the injector heated to 250°C with a split ratio of 10:1. The temperature began at 150°C with a 0.25 min hold, which was followed by a 35°C/min ramp to 200°C, an 8°C/min ramp to 225°C with a 3.2 min hold and finally, an 80°C/min ramp up to 245°C with a 15 min hold. The temperature of the flame ionization detector was 300°C with air and nitrogen make-up gas flow rates of 300 and 25 ml/min, respectively, and a sampling frequency of 50 Hz. An external reference standard (GLC-462, Nu Chek Prep, Elysian, MN) was used to identify individual FA peaks.
In vitro radiochemical triolein hydrolase assay
TAG hydrolase activity was performed as previously described with minor modifications (14, 19). Adipose tissues were homogenized in lysis buffer then centrifuged at 10,000 g to clarify lysates that were adjusted to a final total protein concentration of 1 μg/μl. Samples containing 100 μg protein were added to 100 μl of a reaction mixture containing 300 μM triolein (with 0.15 μCi of [9,10-3H(N)]triolein per reaction), 25 μM egg yolk lecithin, 2% BSA (w/v), 100 μM sodium taurocholate, 50 mM potassium phosphate (pH 7.2), and 1 mM DTT. The reaction mixture was incubated at 37°C for 1 h and quenched by the addition of 0.75 ml of 1:2 chloroform:methanol (v/v). Total lipids were extracted as described by Bligh and Dyer (20) and applied to a silica gel G plate to separate neutral lipids. The band corresponding to hydrolyzed NEFA was identified by comparison with known standards, scraped ,and quantified by liquid scintillation counting.
Statistics
The results are shown as means ± SEM. Significance between two groups was established using Student’s t-test (unpaired, two-tailed) and significance between multiple groups was established by one-way ANOVA with Bonferroni’s posthoc test. Differences were considered significant at P < 0.05.
RESULTS
Agpat4 is expressed in multiple adipose depots but Agpat4 deficiency only alters epididymal WAT weight
Multiple tissues were analyzed for Agpat4 expression in wild-type mice using RT-PCR (Fig. 1A), and Agpat4 expression was also analyzed in several adipose depots, including perirenal, epididymal, retroperitoneal, and inguinal WAT depots, as well as the subscapular brown adipose tissue (BAT) depot (Fig. 1B). Agpat4 expression (relative to 18S) was similar across all adipose tissue depots, and semi-quantitative densitometry analysis indicated no statistically significant differences between different fat pads (Fig. 1B). However, in male Agpat4−/− mice (absence of Agpat4 transcript demonstrated by RT-PCR in epididymal WAT (top) and perirenal WAT (bottom), Fig. 1C), body weights did not differ significantly (Fig. 1D), and no statistically significant differences were seen in weights of perirenal, retroperitoneal, or inguinal WAT or subscapular BAT compared with wild-type littermates (Fig. 1E). Adipose depot weights were expressed relative to total body weight to account for age-related size differences in mice at the time of euthanasia (9–12 weeks of age). Only one significant difference in gross WAT phenotype was evident, and that was a 40% increase in the normalized weight of the epididymal WAT depot in male Agpat4−/− mice versus wild-type control littermates (24.7 ± 2.0 vs. 17.6 ± 1.5 mg tissue/g body weight, P < 0.05). Female mice were observed for whole body weights (Fig. 1F) and weights of perirenal, epididymal, retroperitoneal, and inguinal WAT as well as subscapular BAT (Fig. 1G). However, no statistically significant differences were observed between Agpat4−/− mice and their wild-type counterparts.
Agpat4 gene ablation does not alter energy metabolism, food intake, oxidative substrate utilization, or activity levels
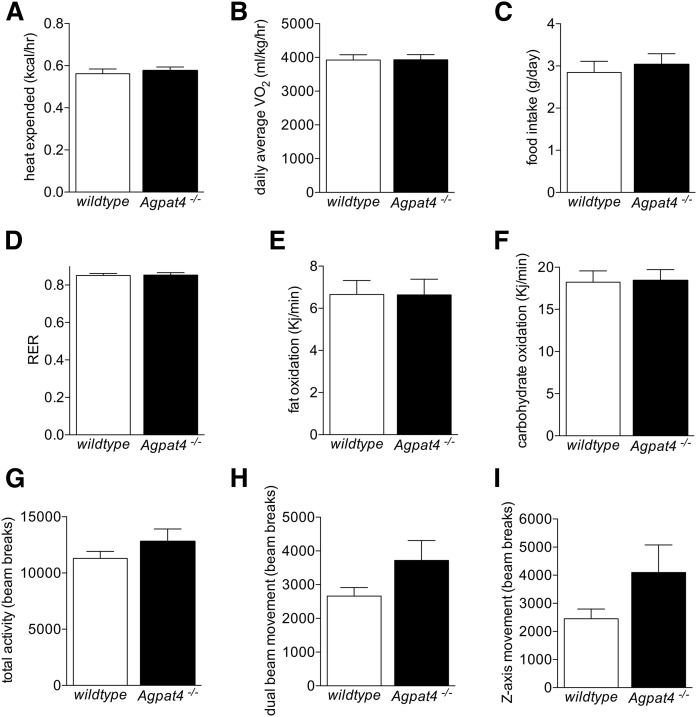
To determine whether the increase in epididymal WAT weight in Agpat4−/− mice is due to differences in energy metabolism, food intake, substrate preference, or activity, mice were housed and continuously monitored for 72 h using the Comprehensive Laboratory Animal Monitoring System. During this time, no significant differences were observed in measures of energy use, including heat expended (Fig. 2A) and daily average VO2 (Fig. 2B). There were also no differences in food intake (Fig. 2C) or fuel substrate preference between Agpat4−/− and wild-type littermate mice, as indicated by measures of the RER (Fig. 2D), fat oxidation (Fig. 2E), and carbohydrate oxidation (Fig. 2F). Activity levels, measured by beam-breaks in X/Y and Z-axes, were also not significantly different between genotypes (Fig. 2G–I).
Fig. 2.
Agpat4 gene ablation does not alter energy metabolism, food intake, oxidative substrate utilization, or activity levels. Measures of energy metabolism (A, B) and food intake (C), relative oxidative substrate utilization (D, E, F), and activity (G, H, I) were determined in male Agpat4−/− mice and wild-type littermates using the Comprehensive Laboratory Animal Monitoring System (n = 14–15). Data are means ± SEM.
TAG and PA are increased in epididymal but not perirenal WAT of Agpat4−/− mice
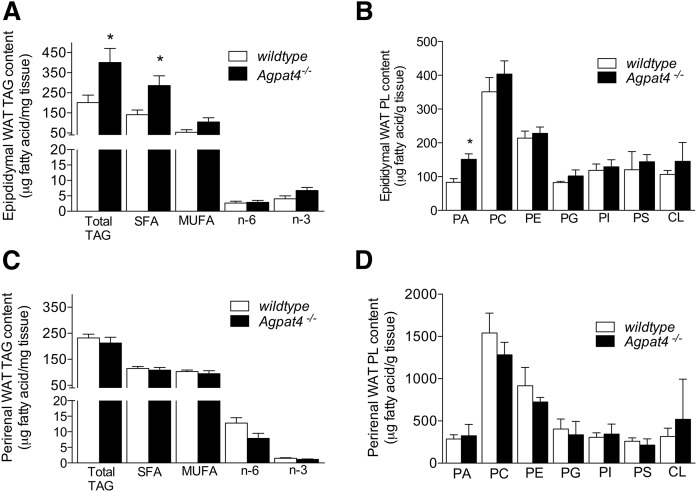
To investigate the nature of the increase in epididymal WAT weights in Agpat4−/− mice, lipid extracts were obtained from this depot and from a second visceral depot (perirenal) for comparison. Epididymal WAT from Agpat4−/− mice had a 2-fold increase in TAG content compared with wild-type mice (400.8 ± 70.3 vs. 201.2 ± 37.0 μg FA/mg adipose tissue, respectively, P < 0.05) (Fig. 3A). This was largely accounted for by a doubling in the content of saturated FAs (286.2 ± 48.3 vs. 141.0 ± 23.2 μg saturated FA/mg, P < 0.05). Differences between wild-type and knockout mice in epididymal TAG MUFA, n-6, or n-3 PUFA contents were not significant (Fig. 3A). Analysis of major phospholipid species indicated that epididymal WAT from Agpat4−/− mice also had a 74% greater total PA content compared with their wild-type littermates (150.8 ± 26.0 vs. 86.5 ± 8.7 μg FA/g adipose tissue, respectively, P < 0.01) (Fig. 3B). However, no further significant differences were observed in the epididymal WAT content of any other class of phospholipid analyzed, including PC, PE, phosphatidylglycerol, PI, phosphatidylserine, or cardiolipin (Fig. 3B). In contrast to findings in epididymal WAT, no significant differences in perirenal WAT were detected between wild-type and knockout mice with regards to the total TAG content or the fatty acyl profile of that TAG (Fig. 3C), or the WAT phospholipid composition (Fig. 3D).
Fig. 3.
TAG and PA content are increased in epididymal but not perirenal WAT of Agpat4−/− mice. Total cellular TAG content and TAG fatty acyl species analysis of male epididymal (A) and perirenal (C) WAT. Phospholipid content of epididymal (B) and perirenal (D) WAT from male wild-type and Agpat4−/− littermate mice. N = 4–5. Data are means ± SEM. *P < 0.05 versus wild-type.
Adipocyte size is increased in epididymal WAT of Agpat4−/− mice but markers of differentiation are not significantly altered
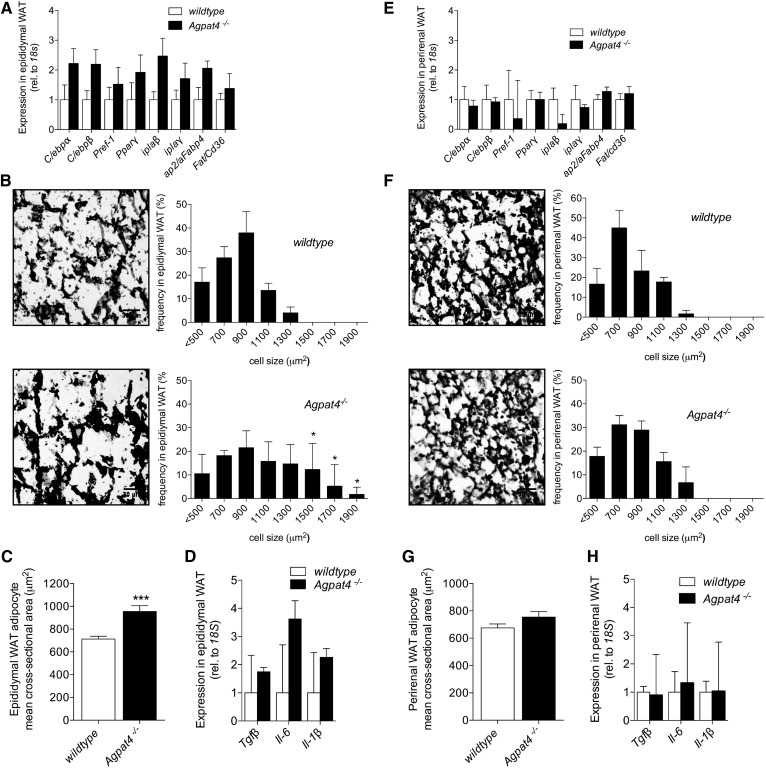
Our observation of elevated TAG concentrations in epididymal WAT from Agpat4−/− mice (Fig. 3A) suggests a hypertrophic rather than a hyperplastic mechanism to explain the increased weight of this depot (Fig. 1D). In agreement, no significant differences were found between epididymal WAT from Agpat4−/− mice and wild-type littermates in the expression of a panel of adipogenesis marker genes including C/ebpα, C/ebpβ, Pref1, Pparγ, iPla2β, iPla2γ, aP2/Fabp4, or Fat/Cd36 (Fig. 4A). Conversely, histological analysis indicated a significantly greater frequency of larger adipocytes in epididymal WAT from Agpat4−/− mice compared with wild-type littermates (Fig. 4B). In particular, adipocytes with a cross-sectional area >1,300 μm2 were evident in epididymal WAT from Agpat4−/− mice that were not evident in wild-type littermates (Fig. 4B), and the mean cross-sectional area of epididymal WAT adipocytes from Agpat4−/− mice was significantly larger (Fig. 4C). No significant differences were found between epididymal WAT from Agpat4−/− mice and wild-type littermates in the gene expression of a panel of inflammatory cytokines (Fig. 4D). As expected, because perirenal WAT depot weights did not differ between Agpat4−/− mice and wild-type littermates, we observed no significant differences between these genotypes with regards to the expression of adipocyte differentiation marker genes (Fig. 4E), the distribution of adipocyte cross-sectional areas (Fig. 4F), mean cell size (Fig. 4G), or in the expression of inflammatory cytokines (Fig. 4H).
Fig. 4.
Agpat−/− mice have larger adipocytes in epididymal but not perirenal WAT compared with wild-type littermates, but expression of differentiation markers is unchanged in either depot. Adipogenic gene expression analysis in epididymal (A) and perirenal (E) WAT (n = 5–7). Representative images of epididymal (B) and perirenal (F) WAT sections, and frequency distribution of adipocyte cell sizes (n = 4). Mean cross-sectional area of adipocytes in epididymal (C) or perirenal (G) WAT sections. Inflammatory cytokine gene expression in epididymal (D) and perirenal (H) WAT (n = 4–5). Data are means ± SEM. *P < 0.05, ***P < 0.001.
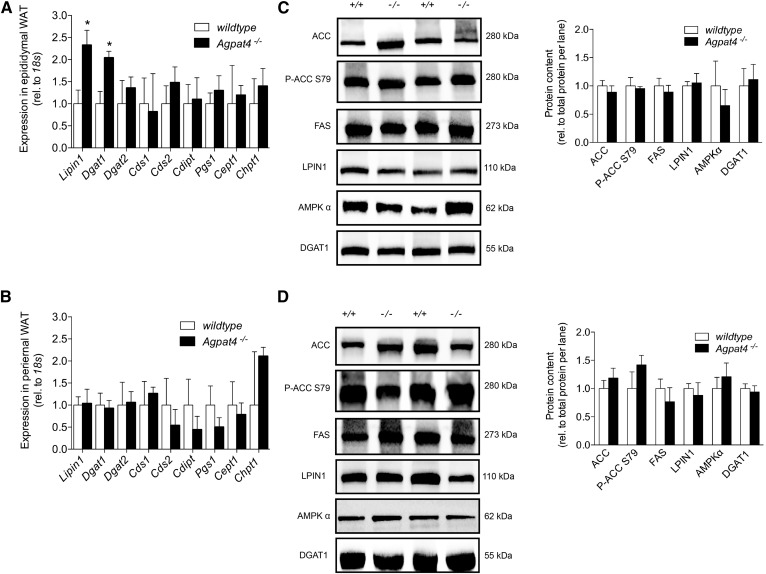
Expression of de novo lipogenesis enzymes and Kennedy pathway enzymes downstream of PA are not significantly altered in Agpat4−/− mice
To assess whether the increase in TAG content and adipocyte size in the epididymal WAT of Agpat4−/− mice may be due to an increase in lipid synthesis, expression of genes encoding Kennedy pathway enzymes was assessed using qPCR (Fig. 5A). Gene analysis of perirenal adipose tissue was also performed (Fig. 5B). There were no significant differences in the expression in epididymal WAT of Dgat2, Cds1, Cds2, Cdipt, Pgs1, Cept1, or Chpt1. In epididymal WAT from Agpat4−/− mice, expression was significantly higher for only two lipid biosynthetic genes, Lpin1 (2.3-fold) and Dgat1 (2.0-fold). However, when protein content was examined by immunoblotting, no significant differences were seen for LPIN1 and DGAT1 (Fig. 5C). No significant differences were seen in the expression of any of these genes in perirenal WAT between Agpat4−/− and wild-type littermates (Fig. 5B). To determine whether an increase in FA synthesis in Agpat4−/− mice may contribute to the observed increase in epididymal TAG mass, immunoblotting was performed on homogenates from knockout mice and wild-type littermates (Fig. 5C). For comparison, perirenal WAT was also analyzed (Fig. 5D). In epididymal WAT, there were no significant differences between Agpat4−/− and wild-type mice in the protein content of FAS, acetyl-CoA carboxylase (ACC), ACC phosphorylated at the deactivating S79 residue, or 5′ AMP-activated protein kinase (AMPK)α that catalyzes this reaction. Similarly, there were no significant differences between mouse genotypes in the levels of any of these proteins in perirenal WAT (Fig. 5D).
Fig. 5.
Expression of de novo lipogenesis enzymes and Kennedy pathway enzymes downstream of PA are not significantly altered in epididymal or perirenal WAT from male Agpat4−/− mice. Anaylsis of a subset of Kennedy pathway genes in epididymal (A) and perirenal (B) WAT (n = 5–7). Representative immunoblots showing content of enzyme regulators of FA de novo biosynthesis and complex lipid synthesis in epididymal (C) and perirenal (D) WAT. Quantification relative to total protein content per lane of immunoblots from epididymal (C, right panel) and perirenal (D, right panel) WATQ6 (n = 4–8). Data are means ± SEM. *P < 0.05 versus wild-type.
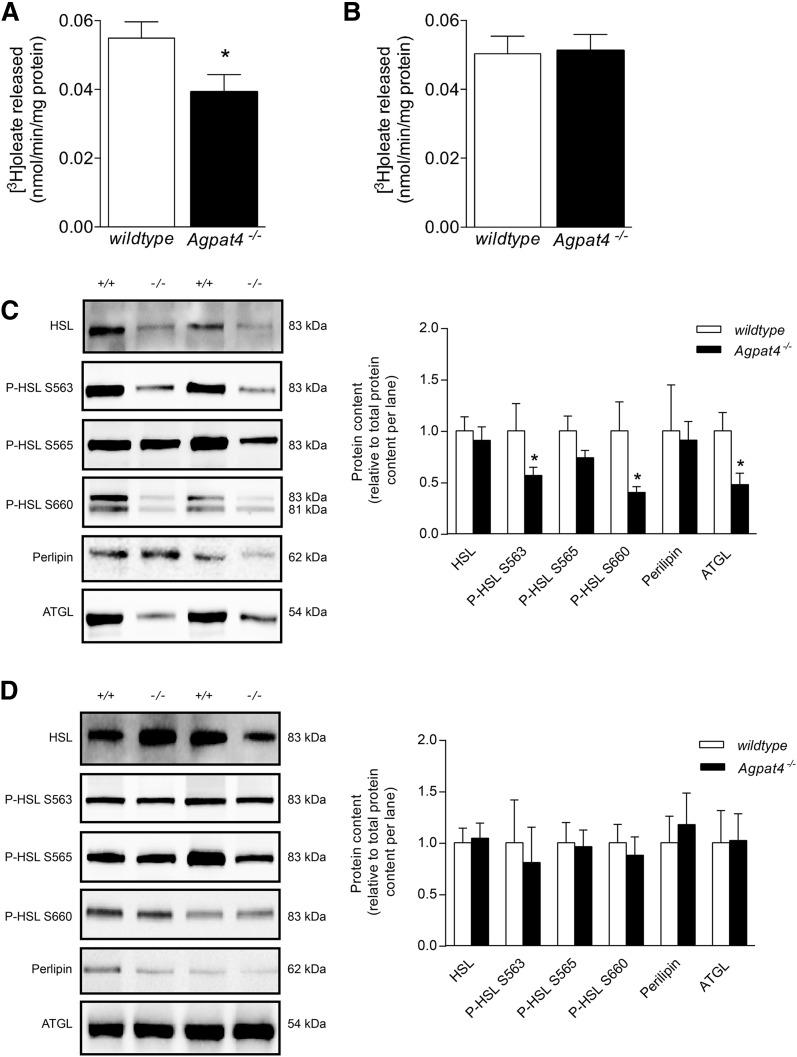
Lipolysis is decreased in epididymal but not perirenal WAT of Agpat4−/− mice
To examine whether the increase in TAG content in the epididymal WAT of Agpat4−/− mice may be related to a reduction in the breakdown of stored fat, measures of lipolysis were performed. First, epididymal (Fig. 6A) and perirenal (Fig. 6B) WAT homogenates from Agpat4−/− and wild-type mice were subjected to a radiochemical [3H]triolein hydrolase assay. Although no differences were observed in perirenal TAG hydrolase activity between genotypes, a significant 29% decrease in [3H]oleic acid liberation was observed from epididymal WAT homogenates of Agpat4−/− mice compared with wild-type littermates (0.039 ± 0.006 vs. 0.055 ± 0.005 nmol/min/mg protein, respectively).
Fig. 6.
Male Agpat4−/− mice have decreased lipolysis in epididymal but not in perirenal WAT. A: Total triolein hydrolase activity of epididymal (A) and perirenal (B) WAT homogenates, determined by assay of the liberation of [3H]oleic acid (n = 6–7). Protein content of lipolytic enzymes and perilipin in epididymal (C) and perirenal (D) WAT, with quantification relative to total protein content per lane (n = 7–9). Data are means ± SEM. *P < 0.05 versus wild-type.
To determine whether the decrease in in vitro lipolytic activity in the epididymal WAT of the Agpat4−/− mice is related to a decrease in the content of enzymes involved in lipolysis, immunoblotting was performed (Fig. 6C). Epididymal WAT from Agpat4−/− mice had significantly less ATGL (52% less), and significantly less phospho-HSL phosphorylated at the protein kinase A-mediated activation sites S563 (42% less) and S660 (60% less), but no significant differences in total HSL or HSL phosphorylated at the S565 AMPKα-site, compared with wild-type littermates. There were also no significant differences in the total epididymal WAT content of perilipin between Agpat4−/− mice and wild-type littermates. Perirenal WAT from Agpat4−/− mice showed no significant differences relative to perirenal WAT from wild-type littermates in content of ATGL, HSL, phospho-HSL S563, phospho-HSL S565, phospho-HSL S660, or perilipin (Fig. 6D).
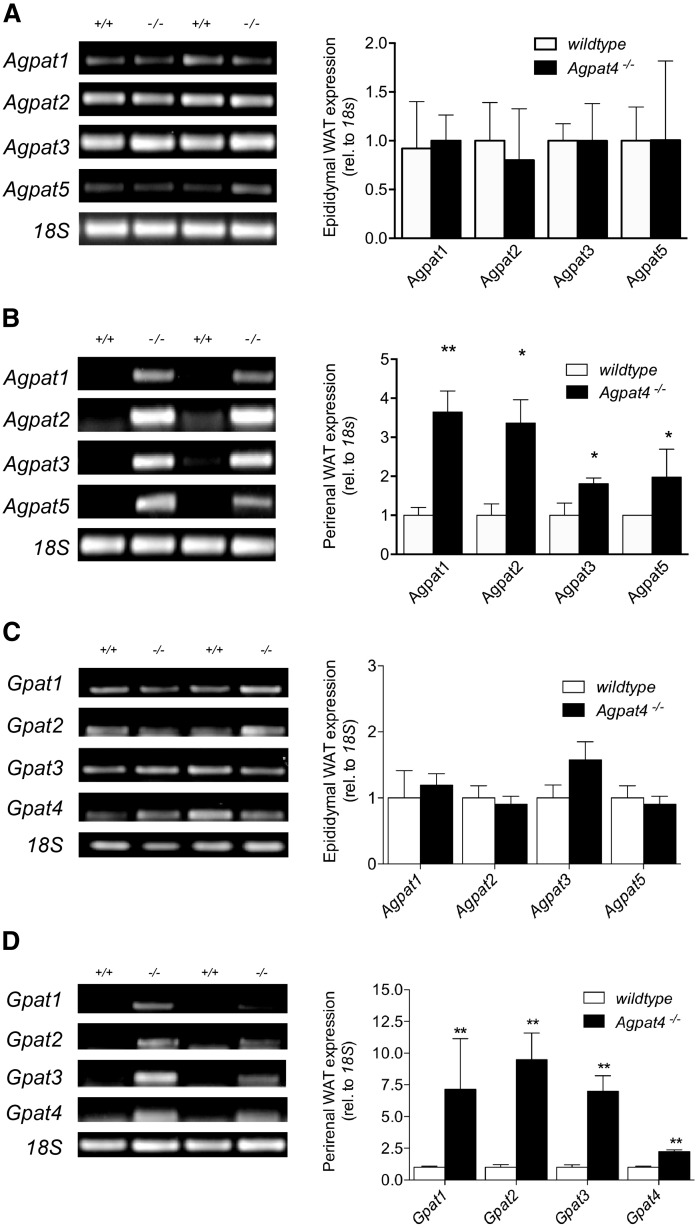
Agpat4−/− mice upregulate alternate Agpat and Gpat homologs in perirenal but not epididymal WAT
Agpat4 is expressed at similar levels in both perirenal and epididymal WAT (Fig. 1B), yet only the latter depot exhibited changes with Agpat4 deficiency in tissue mass, adipocyte size, phospholipid and TAG content, and lipolysis functional measures. To investigate whether differences in the compensatory upregulation of other Agpat and Gpat homologs might help to explain the differential response of perirenal and epididymal WAT to loss of Agpat4, semi-quantitative RT-PCR was performed (Fig. 7). Perirenal WAT from Agpat4−/− mice, which was phenotypically and functionally indistinguishable in our experiments from perirenal WAT of wild-type littermates, showed evidence of a significant induction of all other true Agpats/Lpaats (i.e., Agpat1 (3.65-fold increase), Agpat2 (3.36-fold increase), Agpat3 (1.81-fold increase), and Agpat5 (1.98-fold increase)), as well as all of the known Gpats (i.e., Gpat1 (7.14-fold increase), Gpat2 (9.50-fold increase), Gpat3 (6.99-fold increase), and Gpat4 (2.24-fold increase)), suggesting that a successful compensation for Agpat4 deficiency occurred in this depot (Fig. 7B, D). In contrast, epididymal WAT from Agpat4−/− mice showed no significant upregulation of any other true Agpats/Lpaats or Gpats (Fig. 7A, C), strongly suggesting that alterations in the phenotype and function of epididymal WAT may be related to a failed compensation for Agpat4 deficiency in this depot.
Fig. 7.
Agpat4 deficiency results in upregulation of alternate Agpat and Gpat homologs in perirenal but not epididymal WAT. Representative RT-PCR for Agpat1, Agpat2, Agpat3, and Agpat5 expression, with quantification relative to 18s, in epididymal (A) and perirenal (B) WAT (n = 4). Representative RT-PCR for Gpat1, Gpat2, Gpat3, and Gpat4 expression, with quantification relative to 18S, in epididymal (C) and perirenal (D) WAT (n = 4–5). Data are means ± SEM. *P < 0.05 versus wild-type. **P < 0.01 versus wild-type.
DISCUSSION
The AGPAT family of enzymes contains 11 members, including five true AGPAT/LPAAT isoforms, each showing different tissue expression patterns and substrate preferences (6). We have shown previously that AGPAT4 is highly expressed in the brain (7, 21) and here we demonstrate that it is also abundant in various adipose depots, including epididymal, perirenal, retroperitoneal, and inguinal WAT and subscapular BAT. Other AGPAT family members, including AGPAT2 and AGPAT6, are known to play a significant role in determining adipose TAG content (8, 11, 12). Therefore, we investigated the role of AGPAT4 in WAT using a murine gene ablation model.
Initially, we hypothesized that mice deficient in Agpat4 would exhibit an impaired ability to produce TAG because the AGPAT family of enzymes function in the production of PA, which is a common precursor for both TAG and glycerophospholipid synthesis. As a result, we expected that Agpat4−/− mice would have reduced adiposity and a decrease in whole body mass. Surprisingly, we found that loss of Agpat4 caused no differences in body weight or depot weight in female mice and, indeed, in male mice, caused a significant increase in the weight of the epididymal WAT depot, specifically, without changing whole body weights or causing a significant alteration in weights of the perirenal, retroperitoneal, or inguinal WAT depots or the subscapular BAT depot. These findings are in contrast to Agpat2−/− mice that demonstrate a complete absence of both WAT and BAT, resulting in a redistribution of TAG to the liver and other organs (22). This is also in contrast to Agpat6−/− mice, which have functional adipose tissue depots but exhibit reduced WAT TAG stores, reduced epididymal WAT adipocyte size, and a decrease in total body weights stemming from increased energy expenditure (12). Indeed, compared with the significant metabolic disturbances evident in Agpat2 or Agpat6 knockout mice, Agpat4 knockout mice displayed a milder metabolic phenotype, with no significant changes in energy expenditure, relative substrate oxidation rates, food intake, or activity levels.
We investigated the nature of the difference in epididymal WAT and found that the larger mass of this depot in Agpat4-deficient mice was associated with a greater content of total TAG, resulting primarily from greater stored saturated FAs. The content of PA, which is the direct product of AGPAT4 and a precursor of TAG, was also higher in this tissue. Analysis of the content of the major phospholipid species PC, PE, PI, phosphatidylserine, phosphatidylglycerol, and cardiolipin, as well as expression of a host of phospholipid biosynthesis enzymes, indicated that the change in TAG level was specific and not related to a general increase in synthesis of glycerolipids derived from PA. Expansion of the lipid storage pool suggested that the increase in epididymal WAT mass in Agpat4−/− mice had occurred as a result of an increase in adipocyte size rather than number (23–25). This was confirmed by histochemical analysis, which revealed a greater frequency of larger adipocytes in the epididymal WAT of Agpat4−/− mice and a statistically significant increase in average adipocyte size. Additionally, analysis of epididymal WAT for expression of adipogenic marker genes (e.g., C/ebpα, C/ebpβ, Pref-1, Pparγ, iPlaβ, iPlaγ, Ap2/Fabp4, or Fat/Cd36) indicated no significant differences between genotypes, supporting the conclusion that an increase in adipocyte differentiation (and therefore expansion of cell number) did not contribute significantly to the increased depot mass. For comparison, perirenal WAT depots were also analyzed. As expected, based on the lack of difference in weight of this depot between wild-type and Agpat4−/− mice, no significant differences were observed between genotypes in TAG or phospholipid content, TAG fatty acyl composition, adipocyte size, expression of markers of adipocyte differentiation, or proportion of larger adipocytes.
Lipogenesis and lipolysis are major biochemical functions of adipose tissue that regulate TAG content. Determination of changes in these processes may provide mechanistic insight into the observed increase in TAG in the epididymal WAT of Agpat4−/− mice. No differences were found in epididymal WAT from Agpat4−/− compared with wild-type mice in the content of ACC, P-ACC S79, AMPKα or FAS, which regulate the de novo synthesis of FAs. Upregulation of the mRNA of Lpin1, the gene encoding the enzyme PA phosphatase (26), which catalyzes formation of DAG from PA (27), and Dgat1, which catalyzes the subsequent acylaction of DAG to form TAG (28, 29), was observed in epididymal WAT from Agpat4−/− mice. Because PA has been implicated as a transcriptional regulator of glycerolipid synthesis genes in yeast [reviewed in (30)], it is possible that elevated cellular PA levels may have contributed to induction of Lpin1 and Dgat1. However, this effect of PA has not yet been show to be conserved in mammalian cells and, regardless, when the protein contents of LPIN1 and DGAT1 were examined, no significant differences were observed, indicating that changes in mRNA were not translated to increased enzyme levels and thus were unlikely to have played a role in the increased epididymal TAG observed. The higher PA levels may also have promoted the coalescence of lipid droplets in epididymal WAT, because PA has been found to increase the size of lipid droplets in vitro (31, 32). Further study will be required to determine which possible mechanistic role(s), if any, the higher PA content plays in the observed Agpat4−/− epididymal TAG accumulation. Overall, the data do not support an increase in de novo or complex lipid biosynthesis, and it is possible that the higher PA content in epididymal WAT may reflect inputs from multiple less-conventional sources such as the phosphorylation of DAG or the action of D-type phospholipases. Further investigation using lipidomic and proteomic approaches will be needed to resolve this question. As expected, analysis of perirenal WAT showed no significant changes in depot content of PA or TAG, no changes in levels of FA biosynthetic enzymes, and no changes in any enzymes involved in TAG or glycerophospholipid biosynthesis.
We also explored whether changes in TAG lipolysis could help explain the increase in epididymal WAT weight, adipocyte size, and TAG content observed in Agpat4−/− mice. In a radiochemical assay designed to analyze the total TAG hydrolase activity of adipose tissue homogenates, a significant 30% decrease in FA liberation was observed in homogenates of epididymal WAT from Agpat4−/− mice relative to those of wild-type mice. Adipocyte TAG lipolysis is primarily mediated by the hydrolytic activity of ATGL, which cleaves a fatty acyl chain predominantly from the sn-2 position of TAG forming sn1,3-DAG. HSL subsequently cleaves a fatty acyl chain from either the sn-1 or sn-3 positions of DAG to produce monoacylglycerol, which is hydrolyzed by monoglyceride lipase to liberate the final free FA chain from the glycerol backbone (23). During lipolysis stimulation, intracellular cAMP levels increase, activating cAMP-dependent PKA to phosphorylate HSL at the serine residues 563 and 660, which causes translocation of the enzyme to the lipid droplet (23, 33). Although total HSL levels were unchanged, total ATGL levels were significantly reduced, and decreases in HSL phosphorylated at the activating serine 563 and 660 sites were observed in the epididymal WAT of Agpat4−/− mice. In contrast, no significant differences were observed between genotypes in content of HSL phosphorylated at the AMPKα-activated S565 site. Total perilipin levels also did not differ between genotypes. These findings suggested a decreased capacity for cAMP-stimulated lipolysis in the epididymal WAT of Agpat4−/− mice. It is possible that elevated levels of PA in WAT mediated this effect at least in part, because PA is an allosteric activator of phosphodiesterase 4, which hydrolyzes cAMP to decrease activation of PKA (34, 35). It is also possible that changes in tissues associated with the epididymal fat pad, such as the epididymis and the testes, could interact to influence observed effects of Agpat4 deficiency. Further investigation will be needed to determine the mechanism(s) involved. Similar to results from all other functional and phenotypic measures performed, we found that perirenal WAT depots were indistinguishable between wild-type and Agpat4−/− mice with regards to all measures of lipolytic capacity.
We were somewhat surprised by our findings on the adipose tissue contents of PA. Because AGPAT4 functions to synthesize PA, we had expected a decrease in levels of this phospholipid, but rather found an increase in PA in epididymal WAT and no change in perirenal WAT. These data, however, are not incongruous with prior reports in Agpat knockout models. In male Agpat2 null mice, liver PA content is increased (22). Additionally, we have previously reported that in mouse brain, loss of Agpat4 does not alter total PA levels (7). In that work, it was determined that induction of alternate Agpats/Lpaats likely provided enzymatic compensation that normalized the total tissue PA pool (7). We therefore assessed expression of alternate true Agpat/Lpaat isoforms and Gpat isoforms in epididymal and perirenal WAT. Similar to the effect observed in brain, a compensatory induction of Agpats 1, 2, 3, and 5 as well as Gpats 1, 2, 3, and 4 was observed in perirenal WAT, suggesting that induction of alternate isoforms was sufficient in this depot to restore PA to control levels. However, contrary to results in brain, where levels of the PA-derivatives PC, PE, and PI were decreased despite equivalent total PA contents, we found that the compensated PA level in perirenal WAT resulted in a fully normal complex lipid profile and normal physiological and functional appearance for this depot. Taken together, these findings highlight that PA likely does not form a single pool within cells, but rather is channelled by individual AGPAT/LPAAT homologs into specific substrate pools, supporting the production of different glycerolipids, and this likely occurs in a unique manner in different tissues.
With regards to size, adipocyte characteristics, functionality, and content of major lipid species, perirenal WAT from wild-type and Agpat4−/− littermates was virtually indistinguishable, indicating redundancy of the Agpat4 isoform in this depot. In contrast, compensatory changes in the expression of the other true Agpat/Lpaat isoforms and Gpat isoforms did not occur in epididymal WAT. This indicates a depot-specific difference between perirenal and epididymal WAT in capacity for compensation in response to loss of an Agpat family member. It also indicates that, despite similar levels of Agpat4 expression in both depots, this enzyme may play an essential role in epididymal WAT that is unique from its role in perirenal WAT. And, finally, it suggests that compensation between Agpat as well as Gpat isoforms may be key to maintaining tissue function in some instances, with a failure in this compensation resulting in major perturbations in common cellular processes even when total PA levels do not decrease.
In summary, we have characterized epididymal and perirenal WAT from Agpat4−/− mice and their wild-type littermates and found differences in the biochemistry, molecular physiology, functional processes, and compensatory responses between these two closely positioned visceral depots, despite similar wild-type levels of Agpat4 expression. In perirenal WAT, an induction of the other Agpat/Lpaat isoforms (1, 2, 3, and 5) and Gpat isoforms (1, 2, 3, and 4) appears to have compensated for loss of Agpat4 with regards to restoration of cellular PA levels as well as maintenance of all cellular processes. In epididymal WAT, however, there was no compensatory induction of the other Agpats or Gpats and perturbations were evident in adipocyte size, cellular lipid content, and lipolysis. Findings from this work therefore provide novel information on the differential role of Agpat4 in perirenal and epididymal WAT depots. Findings from this work are also of particular significance for adipose tissue biology research. Differences between visceral and subcutaneous WAT are well characterized, including differences in developmental origin, metabolic activity, mitochondrial content, rate of lipolysis, and correlation to insulin resistance and other metabolic diseases (36–40). The current work, however, indicates a startling heterogeneity in the molecular physiology of closely positioned depots that are often grouped as a single entity (i.e., “visceral adipose tissue”). Our findings therefore highlight the importance of continued investigation to better understand distinguishing characteristics of adipose tissue found in unique subregions of the peritoneal cavity.
Acknowledgments
The authors would like to thank Angela Wagler, Jean Flanagan, and Nancy Gibson and acknowledge their expert assistance in animal care.
Footnotes
Abbreviations:
- ACC
- acetyl-CoA carboxylase
- AGPAT
- acylgylcerophosphate acyltransferase
- AMPK
- AMP-activated protein kinase
- ATGL
- adipose triglyceride lipase
- BAT
- brown adipose tissue
- DAG
- diacylglycerol
- HSL
- hormone-sensitive lipase
- LPA
- lysophosphatidic acid
- LPAAT
- lysophosphatidic acid acyltransferase
- PA
- phosphatidic acid
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PI
- phosphatidylinositol
- RER
- respiratory exchange ratio
- TAG
- triacylglycerol
- VO2
- daily average oxygen consumption
- WAT
- white adipose tissue
This work was supported by infrastructure grants to R.E.D. from the Canada Foundation for Innovation – Leader’s Opportunity Fund and Ontario Research Fund (Project 30259), and by a Natural Sciences and Engineering Research Council (NSERC) of Canada Discovery Grant (418213-2012). K.D.S. is supported through a Canada Research Chair in Nutritional Lipidomics. E.B.M. was supported by an NSERC Canada Graduate Scholarship-Master’s (CGS-M). R.M.B. and J.J.A.H. were supported by NSERC Postgraduate Scholarships-Doctoral (PGS-D).
REFERENCES
- 1.Takeuchi K., and Reue K.. 2009. Biochemistry, physiology, and genetics of GPAT, AGPAT, and lipin enzymes in triglyceride synthesis. Am. J. Physiol. Endocrinol. Metab. 296: E1195–E1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamashita A., Hayashi Y., Matsumoto N., Nemoto-Sasaki Y., Oka S., Tanikawa T., and Sugiura T.. 2014. Glycerophosphate/acylglycerophosphate acyltransferases. Biology (Basel). 3: 801–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kennedy E. P., and Weiss S. B.. 1956. The function of cytidine coenzymes in the biosynthesis of phospholipides. J. Biol. Chem. 222: 193–214. [PubMed] [Google Scholar]
- 4.Prasad S. S., Garg A., and Agarwal A. K.. 2011. Enzymatic activities of the human AGPAT isoform 3 and isoform 5: localization of AGPAT5 to mitochondria. J. Lipid Res. 52: 451–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradley R. M., Mardian E. B., Moes K. A., and Duncan R. E.. 2017. Acute fasting induces expression of acylglycerophosphate acyltransferase (AGPAT) enzymes in murine liver, heart, and brain. Lipids. 52: 457–461. [DOI] [PubMed] [Google Scholar]
- 6.Lu B., Jiang Y. J., Zhou Y., Xu F. Y., Hatch G. M., and Choy P. C.. 2005. Cloning and characterization of murine 1-acyl-sn-glycerol 3-phosphate acyltransferases and their regulation by PPARalpha in murine heart. Biochem. J. 385: 469–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradley R. M., Marvyn P. M., Aristizabal Henao J. J., Mardian E. B., George S., Aucoin M. G., Stark K. D., and Duncan R. E.. 2015. Acylglycerophosphate acyltransferase 4 (AGPAT4) is a mitochondrial lysophosphatidic acid acyltransferase that regulates brain phosphatidylcholine, phosphatidylethanolamine, and phosphatidylinositol levels. Biochim. Biophys. Acta. 1851: 1566–1576. [DOI] [PubMed] [Google Scholar]
- 8.Agarwal A. K., Arioglu E., De Almeida S., Akkoc N., Taylor S. I., Bowcock A. M., Barnes R. I., and Garg A.. 2002. AGPAT2 is mutated in congenital generalized lipodystrophy linked to chromosome 9q34. Nat. Genet. 31: 21–23. [DOI] [PubMed] [Google Scholar]
- 9.Cao J., Li J. L., Li D., Tobin J. F., and Gimeno R. E.. 2006. Molecular identification of microsomal acyl-CoA:glycerol-3-phosphate acyltransferase, a key enzyme in de novo triacylglycerol synthesis. Proc. Natl. Acad. Sci. USA. 103: 19695–19700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao J., Perez S., Goodwin B., Lin Q., Peng H., Qadri A., Zhou Y., Clark R. W., Perreault M., Tobin J. F., et al. . 2014. Mice deleted for GPAT3 have reduced GPAT activity in white adipose tissue and altered energy and cholesterol homeostasis in diet-induced obesity. Am. J. Physiol. Endocrinol. Metab. 306: E1176–E1187. [DOI] [PubMed] [Google Scholar]
- 11.Gale S. E., Frolov A., Han X., Bickel P. E., Cao L., Bowcock A., Schaffer J. E., and Ory D. S.. 2006. A regulatory role for 1-acylglycerol-3-phosphate-O-acyltransferase 2 in adipocyte differentiation. J. Biol. Chem. 281: 11082–11089. [DOI] [PubMed] [Google Scholar]
- 12.Vergnes L., Beigneux A. P., Davis R., Watkins S. M., Young S. G., and Reue K.. 2006. Agpat6 deficiency causes subdermal lipodystrophy and resistance to obesity. J. Lipid Res. 47: 745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bombardier E., Smith I. C., Gamu D., Fajardo V. A., Vigna C., Sayer R. A., Gupta S. C., Bal N. C., Periasamy M., and Tupling A. R.. 2013. Sarcolipin trumps beta-adrenergic receptor signaling as the favored mechanism for muscle-based diet-induced thermogenesis. FASEB J. 27: 3871–3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmadian M., Duncan R. E., Varady K. A., Frasson D., Hellerstein M. K., Birkenfeld A. L., Samuel V. T., Shulman G. I., Wang Y., Kang C., et al. . 2009. Adipose overexpression of desnutrin promotes fatty acid use and attenuates diet-induced obesity. Diabetes. 58: 855–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaworski K., Ahmadian M., Duncan R. E., Sarkadi-Nagy E., Varady K. A., Hellerstein M. K., Lee H. Y., Samuel V. T., Shulman G. I., Kim K. H., et al. . 2009. AdPLA ablation increases lipolysis and prevents obesity induced by high-fat feeding or leptin deficiency. Nat. Med. 15: 159–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider C. A., Rasband W. S., and Eliceiri K. W.. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Folch J., Lees M., and Sloane Stanley G. H.. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226: 497–509. [PubMed] [Google Scholar]
- 18.Metherel A. H., Taha A. Y., Izadi H., and Stark K. D.. 2009. The application of ultrasound energy to increase lipid extraction throughput of solid matrix samples (flaxseed). Prostaglandins Leukot. Essent. Fatty Acids. 81: 417–423. [DOI] [PubMed] [Google Scholar]
- 19.Soni K. G., Lehner R., Metalnikov P., O’Donnell P., Semache M., Gao W., Ashman K., Pshezhetsky A. V., and Mitchell G. A.. 2004. Carboxylesterase 3 (EC 3.1.1.1) is a major adipocyte lipase. J. Biol. Chem. 279: 40683–40689. [DOI] [PubMed] [Google Scholar]
- 20.Bligh E. G., and Dyer W. J.. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 21.Bradley R. M., Mardian E. B., Marvyn P. M., Vasefi M. S., Beazely M. A., Mielke J. G., and Duncan R. E.. 2015. Data on acylglycerophosphate acyltransferase 4 (AGPAT4) during murine embryogenesis and in embryo-derived cultured primary neurons and glia. Data Brief. 6: 28–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cortés V. A., Curtis D. E., Sukumaran S., Shao X., Parameswara V., Rashid S., Smith A. R., Ren J., Esser V., Hammer R. E., et al. . 2009. Molecular mechanisms of hepatic steatosis and insulin resistance in the AGPAT2-deficient mouse model of congenital generalized lipodystrophy. Cell Metab. 9: 165–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duncan R. E., Ahmadian M., Jaworski K., Sarkadi-Nagy E., and Sul H. S.. 2007. Regulation of lipolysis in adipocytes. Annu. Rev. Nutr. 27: 79–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gregoire F. M., Smas C. M., and Sul H. S.. 1998. Understanding adipocyte differentiation. Physiol. Rev. 78: 783–809. [DOI] [PubMed] [Google Scholar]
- 25.Spalding K. L., Arner E., Westermark P. O., Bernard S., Buchholz B. A., Bergmann O., Blomqvist L., Hoffstedt J., Naslund E., Britton T., et al. . 2008. Dynamics of fat cell turnover in humans. Nature. 453: 783–787. [DOI] [PubMed] [Google Scholar]
- 26.Donkor J., Sariahmetoglu M., Dewald J., Brindley D. N., and Reue K.. 2007. Three mammalian lipins act as phosphatidate phosphatases with distinct tissue expression patterns. J. Biol. Chem. 282: 3450–3457. [DOI] [PubMed] [Google Scholar]
- 27.Smith S. W., Weiss S. B., and Kennedy E. P.. 1957. The enzymatic dephosphorylation of phosphatidic acids. J. Biol. Chem. 228: 915–922. [PubMed] [Google Scholar]
- 28.Chen H. C., Stone S. J., Zhou P., Buhman K. K., and Farese R. V. Jr. 2002. Dissociation of obesity and impaired glucose disposal in mice overexpressing acyl coenzyme a:diacylglycerol acyltransferase 1 in white adipose tissue. Diabetes. 51: 3189–3195. [DOI] [PubMed] [Google Scholar]
- 29.Oelkers P., Behari A., Cromley D., Billheimer J. T., and Sturley S. L.. 1998. Characterization of two human genes encoding acyl coenzyme A:cholesterol acyltransferase-related enzymes. J. Biol. Chem. 273: 26765–26771. [DOI] [PubMed] [Google Scholar]
- 30.Carman G. M., and Henry S. A.. 2007. Phosphatidic acid plays a central role in the transcriptional regulation of glycerophospholipid synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 282: 37293–37297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barneda D., Planas-Iglesias J., Gaspar M. L., Mohammadyani D., Prasannan S., Dormann D., Han G. S., Jesch S. A., Carman G. M., Kagan V., et al. . 2015. The brown adipocyte protein CIDEA promotes lipid droplet fusion via a phosphatidic acid-binding amphipathic helix. eLife. 4: e07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fei W., Shui G., Zhang Y., Krahmer N., Ferguson C., Kapterian T. S., Lin R. C., Dawes I. W., Brown A. J., Li P., et al. . 2011. A role for phosphatidic acid in the formation of “supersized” lipid droplets. PLoS Genet. 7: e1002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaworski K., Sarkadi-Nagy E., Duncan R. E., Ahmadian M., and Sul H. S.. 2007. Regulation of triglyceride metabolism. IV. Hormonal regulation of lipolysis in adipose tissue. Am. J. Physiol. Gastrointest. Liver Physiol. 293: G1–G4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitra M. S., Chen Z., Ren H., Harris T. E., Chambers K. T., Hall A. M., Nadra K., Klein S., Chrast R., Su X., et al. . 2013. Mice with an adipocyte-specific lipin 1 separation-of-function allele reveal unexpected roles for phosphatidic acid in metabolic regulation. Proc. Natl. Acad. Sci. USA. 110: 642–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Savany A., Abriat C., Nemoz G., Lagarde M., and Prigent A. F.. 1996. Activation of a cyclic nucleotide phosphodiesterase 4 (PDE4) from rat thymocytes by phosphatidic acid. Cell. Signal. 8: 511–516. [DOI] [PubMed] [Google Scholar]
- 36.Chau Y. Y., Bandiera R., Serrels A., Martinez-Estrada O. M., Qing W., Lee M., Slight J., Thornburn A., Berry R., McHaffie S., et al. . 2014. Visceral and subcutaneous fat have different origins and evidence supports a mesothelial source. Nat. Cell Biol. 16: 367–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deveaud C., Beauvoit B., Salin B., Schaeffer J., and Rigoulet M.. 2004. Regional differences in oxidative capacity of rat white adipose tissue are linked to the mitochondrial content of mature adipocytes. Mol. Cell. Biochem. 267: 157–166. [DOI] [PubMed] [Google Scholar]
- 38.Hajer G. R., van Haeften T. W., and Visseren F. L.. 2008. Adipose tissue dysfunction in obesity, diabetes, and vascular diseases. Eur. Heart J. 29: 2959–2971. [DOI] [PubMed] [Google Scholar]
- 39.Kraunsøe R., Boushel R., Hansen C. N., Schjerling P., Qvortrup K., Støckel M., Mikines K. J., and Dela F.. 2010. Mitochondrial respiration in subcutaneous and visceral adipose tissue from patients with morbid obesity. J. Physiol. 588: 2023–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wajchenberg B. L. 2000. Subcutaneous and visceral adipose tissue: their relation to the metabolic syndrome. Endocr. Rev. 21: 697–738. [DOI] [PubMed] [Google Scholar]