Fig. 5.
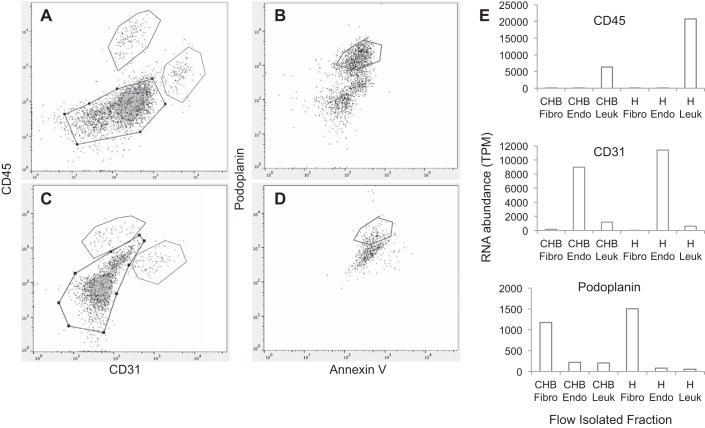
Authentication of enriched leukocytes, endothelial cells, and fibroblasts based on flow sorting and transcriptome analysis. CHB heart (A and B) and control heart (C and D) were stained following the protocols outlined in materials and methods. After the selection of viable (DAPI negative) cells (not shown), anti-CD45 and anti-CD31 were used to separate leukocytes and endothelial cells, respectively (A and C). Note within the same panels the utility of a portion of the unstained cells (gate, black) to further separate isolated cells using anti-podoplanin (B and D). Flow panels show the gating, as indicated on the labels of y- and x-axes of A–D, to isolate CD45, CD31, and podoplanin fractions that are enriched populations of leukocytes, endothelial cells, and fibroblasts (E). As shown in E, RNA abundance of varied isolated cells after flow sorting (which was derived from RNASeq) was consistent with the lineage assignment.