ABSTRACT
Staphylococcus epidermidis has emerged as an important opportunistic pathogen causing orthopedic-device-related infections (ODRI). This study investigated the association of genome variation and phenotypic features of the infecting S. epidermidis isolate with the clinical outcome for the infected patient. S. epidermidis isolates were collected from 104 patients with ODRI. Their clinical outcomes were evaluated, after an average of 26 months, as either “cured” or “not cured.” The isolates were tested for antibiotic susceptibility and biofilm formation. Whole-genome sequencing was performed on all isolates, and genomic variation was related to features associated with “cured” and “not cured.” Strong biofilm formation and aminoglycoside resistance were associated with a “not-cured” outcome (P = 0.031 and P < 0.001, respectively). Based on gene-by-gene analysis, some accessory genes were more prevalent in isolates from the “not-cured” group. These included the biofilm-associated bhp gene, the antiseptic resistance qacA gene, the cassette chromosome recombinase-encoding genes ccrA and ccrB, and the IS256-like transposase gene. This study identifies biofilm formation and antibiotic resistance as associated with poor outcome in S. epidermidis ODRI. Whole-genome sequencing identified specific genes associated with a “not-cured” outcome that should be validated in future studies. (The study has been registered at ClinicalTrials.gov with identifier NCT02640937.)
KEYWORDS: Staphylococcus epidermidis, MRSE, virulence factors, antibiotic resistance, genotype, phenotype, orthopedic-device-related infections
INTRODUCTION
Staphylococcus epidermidis is a common member of the human skin microflora, predominant in moist sites such as nares or fossae and in sebaceous areas such as the facial skin. With the advent of implanted and indwelling medical devices, S. epidermidis has emerged as a prominent cause of nosocomial and device-associated infections (1, 2). The microorganism's ability to switch from a commensal to pathogenic lifestyle is facilitated by its ability to rapidly attach to, and form biofilms upon, medical devices. In the case of orthopedic-device-related infections (ODRI), S. epidermidis accounts for up to 43% of cases and is second only to Staphylococcus aureus as the most prevalent causative organism (1, 2).
Molecular epidemiological studies have begun to reveal information on both the population structure and genetic diversity within S. epidermidis populations (3–5). The complete S. epidermidis genome is estimated at approximately 2.5 Mb and comprises 80% core genes and 20% variable genes (3, 4, 6). Three distinct phylogenetic groups (clades) are evident in the population structure of S. epidermidis (3, 4, 6), with at least nine globally disseminated clonal complex (CC) lineages. The most common clonal complex (CC2) contains one particularly prominent sequence type (ST), ST2 (32% of all isolates) (5, 7).
In an attempt to identify the features that enable invasive infection in S. epidermidis, a number of studies have searched for features that may distinguish invasive from commensal S. epidermidis isolates on genotypic and phenotypic levels. Such studies have identified features such as IS256, the folate dehydrogenase gene, and copper remediation genes to be more common among invasive isolates (3, 6, 8, 9). However, clear separation between the two has proven to be difficult (9–11), perhaps indicating that the ability to invade the host and the ability to colonize it do not require significantly different genetically encoded features. One possibility, which has not been explored to date, however, is whether the genome/phenotype of the invasive isolate dictates the ultimate course of an infection, i.e., whether the patient eventually has a successful treatment outcome or a failed treatment outcome.
In the present study, S. epidermidis isolates were prospectively collected from patients with ODRI and were assigned a clinical outcome (either “cured” or “not cured”) after an extended patient follow-up (FUP). Clinical outcome was then related to genome variation and phenotypes believed to be important for S. epidermidis virulence.
RESULTS
Patient outcome and clinical parameters.
A total of 104 patients with S. epidermidis ODRI were included in this study; complete demographic information is shown in Table 1. The lower-extremity cohort (70 patients) included only those patients with infection of the hip, knee, and upper ankle joints as well as the femur, tibia, and fibula. The majority of patients of the complete cohort study (n = 85, 81.7%) were considered to have had a “cured” clinical outcome at FUP.
TABLE 1.
Patient health status, infection characteristics, bacteriology, clinical course, and outcome
| Parameter | No. (%) of patients |
|
|---|---|---|
| Complete study cohort | Lower-extremity cohort | |
| Totala | 104 (100.0) | 70 (100.0) |
| Clinical course and infection outcome | ||
| Multiple revision surgeries | 89 (85.6) | 68 (97.1) |
| Clinical outcome, cured | 85 (81.7) | 53 (75.7) |
| Health status | ||
| Obesityb | 44 (42.3) | 32 (45.7) |
| Smoking | 28 (26.9) | 17 (24.3) |
| Diabetes | 14 (13.5) | 8 (11.4) |
| Chronic immunosuppression | 25 (24.0) | 14 (20.0) |
| Infection characteristics | ||
| Infection after fracture fixation | 78 (75.0) | 44 (62.9) |
| Prosthetic joint infection | 26 (25.0) | 26 (37.1) |
| Acute infection | 29 (27.9) | 20 (28.6) |
| Closed fracturec | 53 (67.9) | 25 (56.8) |
| Open fracture | 25 (32.1) | 19 (43.2) |
| Type of implant | ||
| Internal fixator | 6 (5.8) | 0 (0) |
| Prosthetic joint | 26 (25.0) | 26 (37.1) |
| Nail | 23 (22.1) | 20 (28.6) |
| Plate | 40 (38.5) | 22 (31.4) |
| Screw | 8 (7.7) | 2 (2.9) |
| K-wire | 1 (1.0) | 0 (0) |
| Localization | ||
| Spine | 6 (5.8) | NAd |
| Upper extremity | 7 (6.7) | NA |
| Pelvis | 7 (6.7) | NA |
| Tibia | 2 (1.9) | NA |
| Clavicle | 3 (2.9) | NA |
| Hip joint | 10 (9.6) | 10 (14.3) |
| Femur | 7 (6.7) | 7 (10.0) |
| Knee joint | 18 (17.3) | 18 (25.7) |
| Lower leg, including upper ankle joint | 35 (33.7) | 35 (50.0) |
| Lower ankle joint, including foot | 9 (8.7) | NA |
| Bacteriological evaluation | ||
| Methicillin resistance | 70 (67.3) | 52 (74.3) |
| Multidrug resistance | 77 (74.0) | 56 (80.0) |
| Biofilm formation | ||
| None | 31 (29.8) | 24 (34.3) |
| Weak | 39 (37.5) | 26 (37.1) |
| Intermediate | 22 (21.2) | 11 (15.7) |
| Strong | 12 (11.5) | 9 (12.9) |
Each patient had 1 S. epidermidis isolate.
Defined as a BMI of >30.
Prosthetic joint infection not included.
NA, not applicable.
Those considered to have a “not-cured” clinical outcome at FUP were statistically more likely to have had multiple revision surgeries in comparison with those with “cured” outcome isolates (P < 0.067) (Table 2). There was no association between outcome and any of the other monitored parameters, such as diabetes, chronic immunosuppression, or obesity (Table 2).
TABLE 2.
Association between prognostic factors and cure status for the complete study cohort
| Parameter | No. (%) of patients in: |
Odds ratio for cureda (95% confidence interval) | P valueb | |
|---|---|---|---|---|
| “Not-cured” group | “Cured” group | |||
| Total no. of patientsc | 19 (18.3) | 85 (81.7) | ||
| Infection type | 0.92 (0.30;2.86) | 1.000† | ||
| FFI | 14 (17.9) | 64 (82.1) | ||
| PJI | 5 (19.2) | 21 (80.8) | ||
| Fracture | 0.39 (0.12;1.27) | 0.126† | ||
| Closed | 7 (13.2) | 46 (86.8) | ||
| Open | 7 (28.0) | 18 (72.0) | ||
| Acute infection | 1.56 (0.44;7.08) | 0.463†† | ||
| No | 15 (20.0) | 60 (80.0) | ||
| Yes | 4 (13.8) | 25 (86.2) | ||
| Multiple revision surgery | 0.12 (0.00;2.04) | 0.067† | ||
| No | 0 (0.0) | 15 (100.0) | ||
| Yes | 19 (21.3) | 70 (78.7) | ||
| Obesity | 0.78 (0.25;2.42) | 0.621†† | ||
| No | 10 (16.7) | 50 (83.3) | ||
| Yes | 9 (20.5) | 35 (79.5) | ||
| Smoking | 0.76 (0.23;2.74) | 0.613†† | ||
| No | 13 (17.1) | 63 (82.9) | ||
| Yes | 6 (21.4) | 22 (78.6) | ||
| Diabetes | 0.50 (0.14;1.81) | 0.281† | ||
| No | 15 (16.7) | 75 (83.3) | ||
| Yes | 4 (28.6) | 10 (71.4) | ||
| Chronic immunosuppression | 0.46 (0.16;1.34) | 0.233† | ||
| No | 12 (15.2) | 67 (84.8) | ||
| Yes | 7 (28.0) | 18 (72.0) | ||
For calculation of odds ratios involving cells with 0 observations, the 0.5 zero-cell correction was applied.
†, chi-square test; ††, Fisher's exact test.
Each patient had 1 S. epidermidis isolate.
Patient outcome and phenotypic properties of isolates. (i) Antibiotic susceptibility.
Antibiotic susceptibility testing of the 104 S. epidermidis isolates found that 74% (77/104) were multiply resistant isolates and 67.3% (70/104) were resistant to methicillin (Table 1). Rifampin resistance was also observed in 19.2% (20/104) of the isolates, which is notable due to the critical role of this antibiotic in treating ODRI. Resistance to aminoglycosides had a statistically significant influence on a “not-cured” clinical outcome (P = 0.001) (Table 3). Further antibiotic resistance (including resistance to aminoglycosides) had no statistically significant influence on any of the other prognostic parameters, such as chronic or acute ODRI. Although isolates from the group of chronic ODRI were more often resistant to aminoglycosides than isolates from the acute-ODRI group (42.7% of versus 31%), this was not statistically significant (P = 0.276). Furthermore, multidrug resistance also showed no statistically significant difference between chronic and acute ODRI (73.3% versus 75.9%).
TABLE 3.
Association between bacterial phenotype and clinical cured status
| Parameter | Complete cohort (n = 104) |
Lower-extremity cohort (n = 70) |
||||||
|---|---|---|---|---|---|---|---|---|
| No. (%) of patients in: |
Odds ratio for cured (95% confidence interval) | P valuea | No. (%) of patients in: |
Odds ratio for cured (95% confidence interval) | P valuea | |||
| “Not-cured” group | “Cured” group | “Not-cured” group | “Cured” group | |||||
| Biofilm formation | 0.059††† | 0.031††† | ||||||
| None | 3 (9.7) | 28 (90.3) | 3 (12.5) | 21 (87.5) | ||||
| Weak | 7 (17.9) | 32 (82.1) | 0.49 (0.08;2.43) | 6 (23.1) | 20 (76.9) | 0.48 (0.07;2.64) | ||
| Intermediate | 5 (22.7) | 17 (77.3) | 0.36 (0.05;2.19) | 4 (36.4) | 7 (63.6) | 0.25 (0.03;1.96) | ||
| Strong | 4 (33.3) | 8 (66.7) | 0.21 (0.03;1.62) | 4 (44.4) | 5 (55.6) | 0.18 (0.02;1.51) | ||
| Antibiotic resistanceb | ||||||||
| Methicillin | 0.33 (0.06;1.29) | 0.082†† | 0.54 (0.14;2.16) | 0.529† | ||||
| No | 3 (8.8) | 31 (91.2) | 3 (16.7) | 15 (83.3) | ||||
| Yes | 16 (22.9) | 54 (77.1) | 14 (26.9) | 38 (73.1) | ||||
| Aminoglycosides | 0.17 (0.04;0.56) | <.001†† | 0.051†† | |||||
| No | 5 (7.9) | 58 (92.1) | 5 (14.3) | 30 (85.7) | 0.32 (0.08;1.17) | |||
| Yes | 14 (34.1) | 27 (65.9) | 12 (34.3) | 23 (65.7) | ||||
†, chi-square test; ††, Fisher's exact test; †††, Cochran-Armitage trend test.
Not all antibiotic resistances are listed. Others tested showed no statistical significance.
(ii) Biofilm formation.
As shown in Table 1, 70.2% (73/104) of the isolates formed a biofilm in vitro. The ability to form biofilm was subdivided into weak biofilm formers (37.5% [39/104] of the isolates), intermediate biofilm formers (21.2%, 22/104), and strong biofilm formers (11.5%, 12/104). The remaining isolates (29.8%, 31/104) were unable to form a biofilm under our in vitro conditions. A statistically significant association between biofilm-forming ability and clinical “cured” versus “not-cured” outcome was noted for the lower-extremity cohort (P = 0.031) (Table 3). Strong biofilm-forming ability also resulted in the highest percentage of “not-cured” outcome for the complete cohort (33.3%, P = 0.059) (Table 3). However, the strength of biofilm formation had no statistically significant influence on any of the prognostic variables, such as multiple revision surgery.
A description of biofilm-associated genes and their relative presence with respect to in vitro biofilm-forming ability is shown in Table 4. Among strongly biofilm-forming isolates, the intercellular adhesion (icaA) gene was more prevalent (83.3%) than the accumulation-associated protein-encoding aap gene (8.3%), the bhp (cell wall-associated biofilm protein) gene (16.7%), and the embP (extracellular matrix-binding protein) gene (66.7%).
TABLE 4.
Biofilm-associated genes and biofilm formation
| Biofilm-associated gene | No. (%) of isolates with gene and the indicated biofilm strength in vitro |
|||||||
|---|---|---|---|---|---|---|---|---|
| Complete cohort study (n = 104) |
Lower-extremity cohort study (n = 70) |
|||||||
| None (n = 31) | Weak (n = 39) | Intermediate (n = 22) | Strong (n = 12) | None (n = 24) | Weak (n = 26) | Intermediate (n = 11) | Strong (n = 9) | |
| icaA | 7 (22.6) | 7 (18.0) | 8 (36.4) | 10 (83.3) | 7 (29.2) | 5 (19.2) | 6 (54.5) | 8 (88.9) |
| aap | 1 (3.2) | 0 (0) | 0 (0) | 1 (8.3) | 1 (4.2) | 0 (0) | 0 (0) | 0 (0) |
| bhp | 7 (22.6) | 10 (25.7) | 0 (0) | 2 (16.7) | 4 (16.7) | 9 (34.6) | 0 (0) | 1 (11.1) |
| embP | 28 (90.3) | 31 (79.5) | 15 (68.2) | 8 (66.7) | 21 (87.5) | 20 (76.9) | 6 (54.5) | 6 (66.7) |
Patient outcome and pathogen genome variation. (i) Relationship between virulence-associated genes and patient outcome.
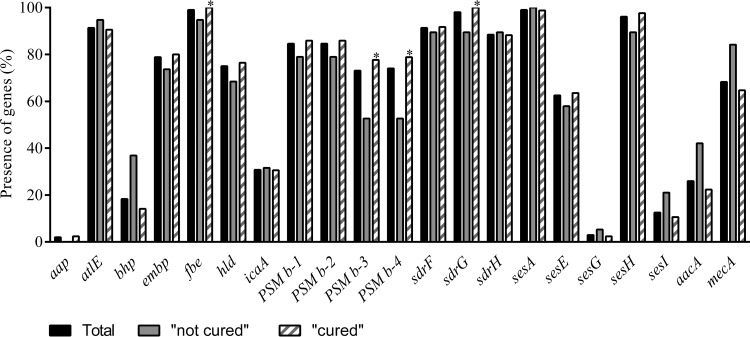
The 104 genomes were analyzed for the presence of a selection of genes previously described as virulence factors in S. epidermidis (Fig. 1) (12–14). Within the population as a whole (i.e., “cured” and “not-cured” outcome isolates), aae (vitronectin), gehC (lipase), gehD (lipase), hlb (β-hemolysin), sesB (S. epidermidis surface protein), and sesC (S. epidermidis surface protein) were present in all 104 isolates. The methicillin resistance gene mecA was carried by 68.3% of the isolates, whereby 69/70 of the phenotypically confirmed methicillin-resistant S. epidermidis (MRSE) isolates and 2/34 of the phenotypically confirmed methicillin-susceptible S. epidermidis (MSSE) isolates possessed this gene. Figure 1 also shows the distribution of the known virulence genes between the 2 clinical outcome groups (“cured” and “not cured”). A trend for the presence of aminoglycoside resistance gene aacA [aac(6′)-aph(2″) and mecA] in the “not-cured” outcome isolates (P = 0.076 and P = 0.099, respectively) was observed. In addition, the presence of biofilm-associated bhp was statistically significantly associated with a “not-cured” clinical outcome in the lower-extremity cohort (P = 0.023).
FIG 1.
Percentages of genes present in the whole collection (black bars) and present in the two outcome groups. Hatched bars, “cured” outcome; gray bars, “not-cured” outcome. *, statistically significant (P ≤ 0.05).
(ii) agr types.
Overall, accessory gene regulator (agr) type I was the most prevalent type (38.5%, 40/104) among the isolates, followed by agr type III (36.5%, 38/104). The distribution of the 3 different agr types within the “cured” and “not-cured” outcome groups are shown in Table 5; however, there were no statistically significant differences (P ≥ 0.05). The only parameter associated with agr type was acute infection (Table 5). All other clinical parameters were not statistically associated with agr type.
TABLE 5.
Association between the agr types and clinical outcome
| Parameter | No. (%) of isolates with agr typea: |
P valueb | ||
|---|---|---|---|---|
| I | II | III | ||
| Clinical outcome | 0.946 | |||
| Not cured | 7 (36.8) | 5 (26.3) | 7 (36.8) | |
| Cured | 33 (39.8) | 19 (22.9) | 31 (37.3) | |
| Infection | 0.002 | |||
| Nonacute (chronic) | 26 (35.1) | 13 (17.6) | 35 (47.3) | |
| Acute | 14 (50.0) | 11 (39.3) | 3 (10.7) | |
Two isolates not belonging to any of the 3 agr groups were excluded for statistical reasons.
Chi-square test.
(iii) MLST.
Within the 104 isolates, 21 different sequence types (STs) were identified based on the 7-locus scheme for S. epidermidis (15) using the build-in multilocus sequence typing (MLST) function of BIGSdb linked with PubMLST databases (4) (Table 6). Thirty isolates could not be assigned to any known ST. While the majority of the ST2 (13/18, 72.2%) and ST5 (16/18, 88.9%) isolates were associated with a “cured” outcome, all ST57 (2/2, 100%), ST89 (1/1, 100%), and S110 (1/1, 100%) isolates were associated with a “not-cured” outcome (Table 6). However, more isolates would be needed to draw statistical conclusions. The identified STs belonged to previously described 7-locus MLST clonal complexes (CCs), of which the largest was CC2 (65/104, 62.5%) (4) (Table 6).
TABLE 6.
MLST of the 104 clinical S. epidermidis isolates
| STa | CCb | No. (%) of isolates |
||
|---|---|---|---|---|
| Total | “Cured” group | “Not-cured” group | ||
| 2 | 2 | 18 (17.3) | 13 (72.2) | 5 (27.8) |
| 5 | 2 | 18 (17.3) | 16 (88.9) | 2 (11.1) |
| 7 | 2 | 1 (1.0) | 1 (100) | 0 (0) |
| 23 | 2 | 4 (3.9) | 4 (100) | 0 (0) |
| 57 | 2 | 2 (1.9) | 0 (0) | 2 (100) |
| 59 | 2 | 6 (5.8) | 5 (83.3) | 1 (16.7) |
| 73 | 2 | 1 (1.0) | 1 (100) | 0 (0) |
| 83 | 2 | 1 (1.0) | 1 (100) | 0 (0) |
| 87 | 2 | 4 (3.9) | 2 (50) | 2 (50) |
| 88 | 2 | 1 (1.0) | 1 (100) | 0 (0) |
| 89 | 2 | 1 (1.0) | 0 (0) | 1 (100) |
| 130 | 2 | 6 (5.8) | 6 (100) | 0 (0) |
| 184 | 2 | 1 (1.0) | 1 (100) | 0 (0) |
| 384 | 2 | 1 (1.0) | 1 (100) | 0 (0) |
| 19 | 147 | 2 (1.9) | 2 (100) | 0 (0) |
| 32 | S32 | 2 (1.9) | 2 (100) | 0 (0) |
| 110 | S110 | 1 (1.0) | 0 (0) | 1 (100) |
| 167 | S167 | 1 (1.0) | 1 (100) | 0 (0) |
| 297 | S297 | 1 (1.0) | 1 (100) | 0 (0) |
| 490 | S490 | 1 (1.0) | 1 (100) | 0 (0) |
| 528 | S528 | 1 (1.0) | 1 (100) | 0 (0) |
| NAc | NA | 30 (28.9) | 25 (83.3) | 5 (16.7) |
| Total | 104 (100) | 85 | 19 | |
Sequence types determined using the built-in MLST function of BIGSdb, linked with PubMLST databases.
Clonal complexes were obtained from previously described data (4).
NA, possible truncation of a corresponding MLST locus at the end of a contig.
Patient outcome and accessory genomes.
Further evaluation of the relative presence of accessory genes that were more prevalent in the “not-cured” outcome group than in the “cured” outcome group is shown in Table 7. S. epidermidis isolates from the “not-cured” outcome group carried the antiseptic resistance-coding gene qacA at a statistically significant higher percentage than isolates from the “cured” outcome group (89.5% versus 27.1%; P = 0.023). Furthermore, the presence of the cassette chromosome recombinase-encoding genes ccrA and ccrB (89.5% versus 23.6% and 89.5% versus 24.7%; P = 0.042 and P = 0.034, respectively) was significantly associated with the “not-cured” isolate genomes.
TABLE 7.
Relative overrepresentation (>20% difference) of accessory genes in the “not-cured” outcome isolates
| Locus | Description of product | “Not cured” group (n = 19) |
“Cured” group (n = 85) |
Difference (%) between “not cured” and “cured” groups | P value | ||
|---|---|---|---|---|---|---|---|
| No. of isolates | Prevalence (%) | No. of isolates | Prevalence (%) | ||||
| SERP0915 | IS256-like transposase | 11 | 57.9 | 31 | 36.5 | 21.4 | 0.085 |
| SERP1222 | Transposase | 9 | 47.4 | 16 | 18.8 | 28.5 | 0.008 |
| SERP1586 | Acetyltransferase, GNAT family | 9 | 47.4 | 22 | 25.8 | 21.5 | 0.064 |
| SERP2498 | Cassette chromosome recombinase A (CcrA) | 17 | 89.5 | 56 | 65.8 | 23.6 | 0.042 |
| SERP2499 | Cassette chromosome recombinase B (CcrB) | 17 | 89.5 | 55 | 64.7 | 24.7 | 0.034 |
| id1043_1239 | Hypothetical protein | 10 | 52.6 | 25 | 29.4 | 23.3 | 0.053 |
| id1044_0888 | Phage protein | 8 | 42.1 | 15 | 17.6 | 24.5 | 0.02 |
| id1044_0895 | Phage antirepressor protein | 8 | 42.1 | 17 | 20 | 22.1 | 0.041 |
| id1044_1909 | Antiseptic resistance protein QacA | 17 | 89.5 | 53 | 62.4 | 27.1 | 0.023 |
| id1044_2610 | Unknown | 8 | 42.1 | 15 | 17.6 | 24.5 | 0.02 |
| id1048_0369 | Replication-associated protein | 11 | 57.9 | 29 | 34.1 | 23.7 | 0.054 |
| id1632_0817 | Zn-dependent hydroxyacylglutathione hydrolase | 14 | 73.7 | 44 | 51.7 | 21.9 | 0.082 |
Core and accessory genome analysis.
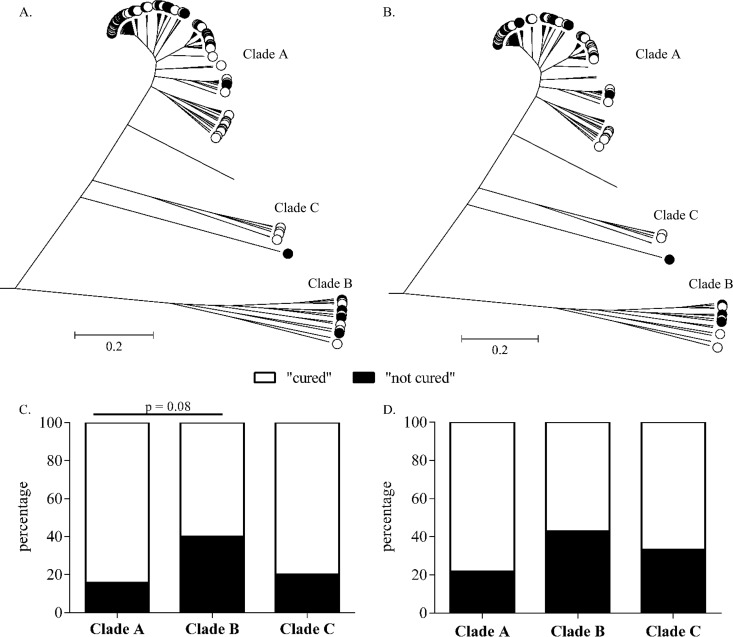
A pan-genome of all study isolates was used to compare the genomes of the 104 clinical S. epidermidis isolates. ClonalFrame was used to construct ancestral genealogies, free from recombination. In order to run ClonalFrame, a stringent approach to selecting core genes based on presence in 100% of the 104 isolates was applied. This resulted in a reduced core genome consisting of 123 nontruncated genes. S. epidermidis isolates clustered into 3 clades (Fig. 2), with 86% of isolates (89/104) in clade A, 9.6% (10/104 isolates) in clade B, and 4.8% (5/104 isolates) in clade C (Fig. 2A).
FIG 2.
Population structure of S. epidermidis isolates constructed from 123 core genes and implemented in ClonalFrame. (A and B) All 104 isolates of the complete cohort (A) and all 70 isolates of the lower-extremity cohort (B) are labeled according to the clinical follow-up (FUP) outcome: “not cured” (black circles) or “cured” (white circles). (C and D) Percent distributions of “cured” and “not-cured” outcome in the three clades A, B, and C, showing the complete cohort (C) and the lower extremity cohort (D).
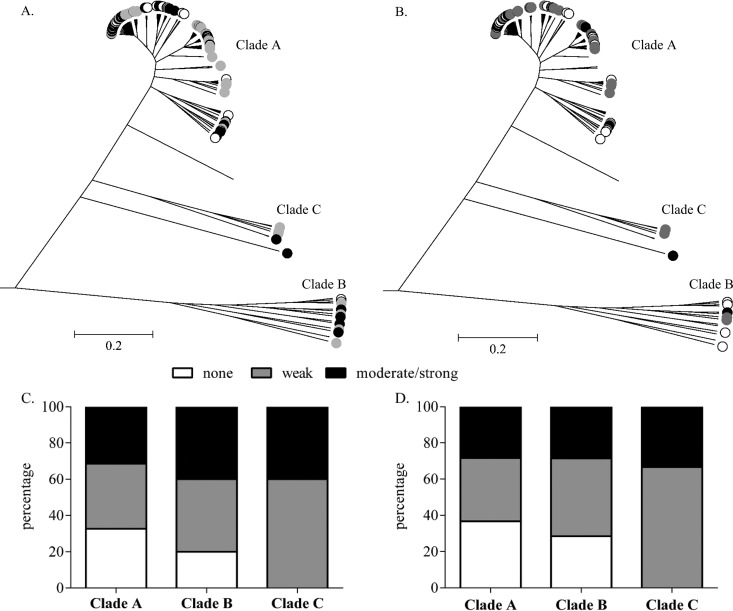
Comparing patient outcome between the clades, a trend between clades A and B was observed. Clade B consisted of a comparatively higher percentage of “not-cured” outcome isolates (40%, 4/10) than clade A (15.7%, 14/89) (Fig. 2A and C). However, this trend did not quite reach statistical significance (P = 0.08; Fisher's exact test). Only 1/5 (20%) clade C isolates belonged to the “not-cured” outcome group, although the low numbers of isolates precluded reliable statistical analysis (Fig. 2B and D). Furthermore, clade B isolates also contained a higher percentage of moderately/strongly biofilm-forming isolates than clade A isolates (40% versus 31.5%) (Fig. 3A and C). In addition, the majority of clade B isolates (80%) belonged to CC2, while clade A possessed only 60.7% CC2 isolates (data not shown).
FIG 3.
Population structure of S. epidermidis isolates constructed from 123 core genes and implemented in ClonalFrame. (A and B) All 104 isolates of the complete cohort (A) and of the lower-extremity cohort (B) are labeled according to the ability to form a biofilm: biofilm negative (white circles), weak biofilm formers (gray circles), and moderate to strong biofilm formers (black circles). (C and D) Percent distribution of the strength of biofilm formation in the three clades A, B, and C, showing the complete cohort (C) and the lower-extremity cohort (D).
Clinical outcome, biofilm formation, and antibiotic resistance phenotypes were homogeneous compared to the clonal frame (P ≥ 0.05). In addition, the permutation test revealed a strong association between lineage and biofilm formation (P ≤ 0.0001) and resistance to methicillin (P = 0.0002), quinolones (P = 0.0055), erythromycin (P < 0.00001), clindamycin (P < 0.00001), tetracycline (P < 0.00001), trimethoprim-sulfonamide (P = 0.02), and fusidic acid (P < 0.00001). However, there was no association between lineage and outcome (P = 0.09) or resistance to penicillin (P = 1), aminoglycosides (P = 0.3798), fosfomycin (P = 0.053), and rifampin (P = 0.151).
DISCUSSION
S. epidermidis is a commensal microorganism that is also a frequent agent of ODRI (6, 8, 16, 17). However, little is known about the impact that genotypic and phenotypic features of the infecting pathogen can have on treatment outcome (3, 6, 8, 9). This prospective study was designed to test the hypothesis that treatment outcome in patients with S. epidermidis ODRI may be influenced by phenotypic or genotypic features of the infecting pathogen. Against a background of scientific studies searching for features that distinguish commensal from invasive isolates (3, 6, 8–11), or for host factors that have an influence on patient outcome (8, 18), this study advances this line of investigation by looking for bacteria-retained features that distinguish infections that result in poor treatment outcome. After prospectively collecting 104 patients, with an average 2-year follow-up (FUP), and subjecting infecting pathogens to genome sequencing and a number of phenotypic assays, we have identified a number of features associated with poor treatment outcome. Those features include biofilm formation, aminoglycoside resistance, the cassette chromosome recombinase encoding genes ccrA and ccrB, IS256-like and plasmid-borne qacA gene, as well as the biofilm-associated bhp gene.
Adhesion to and biofilm formation upon biomaterial substrates are widely believed to be the primary virulence factor enabling invasive S. epidermidis ODRI (6, 8, 16, 17, 19). The data from our study supports this by revealing that a “not-cured” clinical outcome was significantly associated with an increased ability to form biofilm in vitro (P = 0.031). Genomic analysis on the known biofilm-associated genes, such as icaA, aap, bhp, or embP, revealed that the only such gene found to be significantly associated with a “not-cured” outcome was bhp in the lower-extremity cohort (P = 0.023). Interestingly, bhp was most prevalent in the weakly biofilm-forming isolates (52.7%) indicating that its role may not be directly linked with biofilm-forming ability, at least in vitro. bhp has been reported to promote primary attachment to abiotic surfaces as well as intercellular adhesion during biofilm formation (20, 21). Thus, this protein might be important for rapid attachment to the implant rather than the amount of biofilm formed by the isolate per se. A rapid attachment clearly may be significant for early establishment of biofilm in vivo in “the race for the surface.” This may partially explain its association with poor treatment outcome, despite the lack of association with in vitro biofilm-forming ability.
Antibiotic resistance is a second key challenge in treatment of ODRI. Previous studies have suggested that methicillin resistance is associated with a worse treatment outcome in staphylococcal ODRI (22–24), although a number of studies have provided contrasting findings (8, 25, 26). Methicillin resistance is due to the mecA gene. In the present study, resistance to methicillin showed a trend for a “not-cured” patient outcome (P = 0.082 based on phenotypic analysis and P = 0.099 for presence of the mecA gene), supporting previously reported trends (22–24). Furthermore, the chromosome recombinase A- and B-encoding genes ccrB and ccrA were significantly more prevalent in “not-cured” clinical-outcome isolates (89.5% versus 65.8% and 89.5% versus 64.7%; P = 0.042 and P = 0.034, respectively). These two genes are responsible for the chromosomal insertion of the genetic element called staphylococcal cassette chromosome mec (SCCmec). The SCCmec mobile genetic island contains the mec gene complex, including the methicillin resistance gene mecA. In this study, 91.8% of the ccrA/ccrB-positive isolates possessed the mecA gene, indicative for the presence of the mobile element SCCmec. Of those 67 ccrA/ccrB/mecA-positive isolates, only 2 were not phenotypically resistant to methicillin, which might be due to mutations in the mecA gene. The 6 ccrA/ccrB-positive but mecA-negative isolates were not phenotypically resistant to methicillin, indicative of an absent SCCmec mobile element.
A second antibiotic class pertinent to the treatment of ODRI is the aminoglycosides (including gentamicin and tobramycin), which are commonly used in antibiotic-loaded bone cement (2, 16, 19, 27). Resistance to aminoglycosides in S. epidermidis isolated from patients with ODRI typically ranges from 40 to 65% (19, 27). In this study, 39.4% of the isolates were resistant to gentamicin/aminoglycoside, and we observed an association between the “not-cured” outcome and isolates being phenotypically resistant to aminoglycosides (P = 0.001). The majority of the aminoglycoside-resistant isolates (65.8%) carried the aacA [aac(6′)-aph(2″)] gene, which confers resistance to all aminoglycosides. This gene was also observed in a higher prevalence in the “not-cured” group (42.1%, versus 22.4% in “cured”; P = 0.076). This correlates well with other studies in terms of prevalence of the aacA [aac(6′)-aph(2″)] gene among aminoglycoside-resistant isolates, ranging between 40 and 92% (16, 28, 29), although how this impacted treatment outcome was not described in these other studies.
Our data also revealed that the antiseptic (quaternary ammonium compound) qacA gene was statistically more prevalent in the “not-cured” outcome group (P = 0.023). The qacA gene is a plasmid-borne gene (pSK1 family plasmids) that confers resistance to antiseptics and disinfectants such as cetrimide, benzalkonium chloride, and chlorhexidine (30–32). Our observation that the qacA gene was present in 67.3% of isolates (89.5% of “not-cured” group isolates) within the complete cohort seems enriched compared to that in other studies for clinical (47% to 52%) and commensal (25%) S. epidermidis isolates (31, 32). Despite the relatively high presence in our collection and a moderate number of “not-cured” isolates, there is some statistical significance to associate the presence of this gene with a poor treatment outcome. Qac proteins are efflux pumps that protect bacteria not only from a variety of toxic substances but also from fluoroquinolones and β-lactams (30–32). The acquisition of such a gene/plasmid, possibly from antiseptic usage within the hospital, clearly provides the bacteria a survival advantage, especially in a clinical environment. Such resistant pathogens therefore not only are more difficult to clean within the hospital environment but, as we show, also are associated with a poor treatment outcome.
In addition, the IS256-like transposase was more frequently present in “not-cured” clinical-outcome isolates than in “cured” outcome isolates (57.9% versus 36.5%; P = 0.085). Previous studies have described an association between the presence of the IS256 element, the aac(6′)-aph(2[dprime]) gene (33, 34), the icaADBC operon, and the ability to form biofilm (11, 35, 36). Furthermore, IS256 has been suggested as a molecular marker for the molecular typing and identification of nosocomial, invasive S. epidermidis isolates (9–11, 36). This study provides further evidence that IS256 not only is “enriched” within invasive isolates but is also more prevalent in isolates with a poor treatment outcome. The increased prevalence in the “not-cured” group indicates that it is not a marker for infection but rather potentially is one for poor outcome; however, this warrants further study with a larger set of isolates.
Previous genealogical reconstruction studies of S. epidermidis have shown that isolates clustered into 3 phylogenetic clades (4, 6), which is consistent with the observation in this study. To date, no study has associated genotypes with clinical outcomes in ODRI. In this study, a higher number of “not-cured” outcome isolates were found in clade B than in clades A and C. Clade B was also the lineage with the strongest biofilm-forming isolates. Harris et al. reported thick biofilm being 20% more common in CC2 isolates (6). In the present study, CC2 accounted for 80% of the 10 clade B isolates, with 50% of them being responsible for moderate/strong biofilm formation. Furthermore, of these moderately/strongly biofilm-forming CC2 clade B isolates, 75% (3/4) resulted in a “not-cured” outcome. This emphasizes that clade B CC2 isolates might be more likely to result in a poor clinical outcome. However, a greater number of isolates should be analyzed in a prospective manner in order to confirm this observation and determine whether it may be a prognostic molecular marker for poor treatment outcome.
A limitation of this study was that only a single S. epidermidis colony from each patient was analyzed, although the infection could, at least in theory, be polyclonal. A previous study has shown that only a minority of prosthetic joint infections (PJIs) (28.5% [4/14 patients]) were due to polyclonal S. epidermidis strains (37). Any future studies should consider analyzing several colonies from each patient. Furthermore, the morphology of colonies was not recorded in the present study, and so we do not know how many, if any, small-colony variants (SCVs) were present in the current collection, but this should be considered in future studies.
In general, SCVs present phenotypic features such as low growth rate and small colony morphology (38–45). Additionally, SCVs are associated with increased biofilm-forming ability, antibiotic resistance, and ability to internalize and persist in osteoblasts, all of which may contribute to prolonged treatment or even treatment failure (38–45). In contrast to the case for S. aureus SCVs, very little information is available on S. epidermidis SCVs (38, 46). Only recently has the pathogenesis of PJIs been associated with S. epidermidis SCVs (38, 46, 47). Furthermore, SCV colonies from the same patient showed difference in growth rate, colony size, and levels of gentamicin resistance compared to each other (38). This highlights the importance of documenting and analyzing SCVs, as they may influence treatment outcome.
Patients with a “not-cured” clinical outcome were more likely to have had multiple revision surgeries than those with a with “cured” outcome (P < 0.067), which is to be expected as revision surgery is a standard intervention for failed treatment We have considered the final outcome to be “cured” or “not cured” at follow-up, regardless of the treatment steps taken in the interim period. Therefore, even though multiple revision surgeries occurred, if the patient was free of infection at FUP, we considered the patient to be cured. Of course, the need for multiple revisions is possibly an indicator that the infection was a greater challenge to treat; however, in a large patient population such as this, there is often a need for multiple revision surgeries to advance the healing of the fracture or replace the device, which may occur after infection has cleared, and so such patients have also had multiple surgeries.
Overall, genome sequencing is not absolutely required to determine some of the features identified in this study as being associated with poor outcome. For example, routine antibiotic susceptibility testing and conventional in vitro biofilm assays are readily available to provide this information. Nevertheless, whole-genome sequencing allowed us to test our hypothesis with greatest sensitivity and also identified features that are less easily measurable in a clinical laboratory. Finally, it should be mentioned that the treatment of ODRI is achieved by antibiotic therapy and surgical removal of infected tissue. Therefore, the outcome of ODRI treatment will be influenced by these factors in addition to the host defenses and not solely by the pathogen itself. The factors identified in this study therefore require prospective validation in further studies with larger patient cohorts in order to confirm their value as prognostic markers for ODRI treatment outcome.
MATERIALS AND METHODS
Ethics statement.
Institutional Review Board approval was obtained from the local ethical committee Ethik-Kommission der Bayerischen Landesärztekammer under approval number 12063. The study was registered with https://clinicaltrials.gov with identifier NCT02640937. Only adult patients (>18 years) were included in this study, and all patients provided informed written consent prior to inclusion in the study.
Staphylococcus epidermidis collection.
This was a prospective study performed between November 2011 and September 2013 at the BGU Murnau, Germany, a level-one trauma center with a high volume, 70-bed unit for septic and reconstructive surgery. The phenotypic investigation of biofilm formation by a subgroup of these isolates has been previously described (8), although no genome sequence data for these isolates have been previously published.
Inclusion criteria were treatment for a confirmed S. epidermidis infection involving fracture fixation (FFI) or prosthetic joint infections (PJIs). Most of the primary surgeries for fracture fixation or implantation of an endoprosthesis were performed in other hospitals. In cases where the patient developed an infection that was not treated/treatable at the primary center, the patient was transferred to the study site which has a specialized unit for ODRI treatment. Bacterial growth in at least two biopsy specimens collected at the site of interest in combination with nonunion, implant loosening/failure, or local and systemic signs suggesting a surgical site infection were requirements for the diagnosis of ODRI, as per hospital standards.
In the previously described clinical study, patient data were analyzed as a complete study cohort but also as a cohort including only patients with infections associated with the lower limb (8). This is because there are numerous outcome measures for the lower extremity that are not available for other anatomical locations. These outcome measures include the lower-extremity functional score (LEFS) and the short form 12 (SF-12) score, as well as leg length discrepancy (8). The remaining patients, not included in the lower-extremity cohort, included patients with infections at other locations, such as upper extremity, pelvis, and spine (Table 1). At the first surgical procedure after enrollment, bone biopsy specimens were taken from the interface between implant and affected bone. Samples were placed in a sterile container with thioglycolate liquid medium (bioMérieux, Hazelwood, MO, USA) and cultured for 10 days at 37°C. Any growth was inoculated onto a blood agar plate (bioMérieux, Hazelwood, MO, USA) for further growth and subsequent identification. All isolates were grown on tryptone soy agar (TSA) (Oxoid, Pratteln, Switzerland) and incubated overnight at 37°C. A single colony was then taken and resuspended in 1 ml tryptone soy broth (TSB) (Oxoid, Pratteln, Switzerland) containing 20% (vol/vol) glycerol for long-term storage at −80°C. Although the colony morphology of culture-positive samples was not described, we anticipate that SCV colonies had sufficient time to emerge under standard laboratory conditions and are not likely to have been missed in the clinical routine.
Clinical data collection.
Clinical data were collected from each enrolled patient. The following surgical parameters were documented: affected bone or joint, type of implant, time between implantation of the device and onset of symptoms, and whether the fracture was open or closed (PJIs excluded).
Patients were assessed for treatment outcome after an average of 26 months of follow-up (FUP). Patients were assigned to have had a “cured” or a “not-cured” outcome at FUP. Patients had a “cured” clinical outcome if they were free of infection, surgical therapy and systemic antibiotic therapy had ceased, and function of the affected joint or limb was restored. Patients were considered to have had a “not-cured” clinical outcome if at least one of the above parameters was negative. Additional parameters were documented, such as acute/nonacute (chronic) infection (cutoff for onset of symptoms, 6 weeks), obesity (body mass index [BMI] of ≥30 kg/m2), diabetes, smoking, chronic immunosuppressive conditions (diabetes mellitus, chronic alcoholism, Child's class C cirrhosis, neoplasia, transplantation, AIDS, and steroid medication), and whether multiple revision surgeries were required during treatment.
The clinical treatment strategies applied to these patients followed recent guidelines and recommendations, including guidance on antimicrobial stewardship. Therefore, treatment strategies differed between enrolled patients due to antibiotic resistance patterns, presence of implant (yes/no), and stage of treatment. The use of antibiotic-loaded bone cement was not extracted from the patient records; however, in all cases of infection with a gentamicin-resistant organism, any bone cement would have been loaded with vancomycin as the preferred alternative. Whether an implant was removed or retained was dependent upon the classification of the infection and the health status of the patient. In chronic infections, the implant was routinely removed in the first revision surgery whenever possible. In general, the implant was retained in acute infections if sufficient debridement was possible.
Antibiotic susceptibility testing.
Susceptibility to 28 antibiotics was determined using a Vitek2 machine (bioMérieux Vitek Inc., Hazelwood, MO, USA). The antibiotics tested were amikacin, ampicillin-sulbactam, cefotaxime, cefoxitin, cefuroxime, ciprofloxacin, clindamycin, daptomycin, erythromycin, fosfomycin, fusidic acid, gentamicin, levofloxacin, linezolid, mezlocillin, moxifloxacin, netilmicin, ofloxacin, oxacillin, penicillin, piperacillin, rifampin, tetracycline, ticarcillin-clavulanate, tigecycline, tobramycin, trimethoprim-sulfamethoxazole, and vancomycin. Multiple antibiotic resistance was defined according to the definitions of the European Committee of Antimicrobial Susceptibility Testing (EUCAST). Oxacillin resistance was considered definitive for methicillin resistance status.
Biofilm formation.
Biofilm formation was assayed as described previously (48, 49). Briefly, overnight cultures were grown in TSB and then subcultured in fresh TSB containing 1% glucose to approximately 1 × 106 CFU/ml. To achieve this, the bacterial density was adjusted to a target optical density (OD) of known concentration using a Multiskan Go microplate reader (Thermo Scientific, Zürich, Switzerland). A total of 200 μl of the bacterial suspension was incubated in flat-bottom 96-well tissue culture-treated polystyrene microtiter plates (Nuclon; Nunc A/S, Denmark) for 24 h at 37°C. Plates were rinsed with phosphate-buffered saline (PBS) (Sigma-Aldrich, Buchs, Switzerland) and stained with 150 μl of Gram's crystal violet solution (Sigma-Aldrich, Buchs, Switzerland). The dye bound to the attached cells was solubilized by addition of 150 μl of 95% ethanol. Optical density was measured as absorbance at 595 nm using the Multiskan Go microplate reader.
All isolates were tested in triplicate in three independent experiments. Each microtiter plate also consisted of negative controls (wells without bacterial inoculation). The average OD value (ODa) was calculated for each isolate and the negative control. The results were evaluated using the scale described by Stepanovic et al. (49), whereby isolates may fall into the following four categories: biofilm nonproducer, weak biofilm producer, intermediate biofilm producer, and strong biofilm producer. Based on the ODa values and the cutoff value (ODc), which is defined as three standard deviations (SD) above the mean OD of the negative control, ODc = average OD of negative control + (3 × SD of negative control). The strength of the biofilm production of each isolate was calculated as follows: ODa ≤ ODc = biofilm nonproducer, ODc < ODa ≤ 2 × ODc = weak biofilm producer, 2 × ODc < ODa ≤ 4 × ODc = intermediate biofilm producer, and 4 × ODc < ODa = strong biofilm producer. S. epidermidis reference strain RP12 (ATCC 35983) was used as a control for strong biofilm production.
Genome sequencing and assembly.
S. epidermidis isolates were cultured on TSA plates at 37°C for 24 h. Single-colony cultures were harvested, resuspended in 3 ml of TSB medium to minimize clumping, and incubated at 37°C with overnight shaking. Chromosomal DNA was extracted using a Qiagen QIAamp DNA minikit (Qiagen, Hilden, Germany) following the manufacturer's protocol, using 1 μg/ml lysostaphin (Sigma-Aldrich, Buchs, Switzerland) and 2 μg/ml lysozyme (Sigma-Aldrich, Buchs, Switzerland) to facilitate cell lysis. DNA was sequenced at the Swansea University Genome Centre using a MiSeq benchtop sequencer (Illumina, San Diego, CA, USA). Sequencing libraries were prepared using Nextera XT library preparation kits (v2) and paired-end 250-bp reads generated with the MiSeq run kit (v2). Short-read data were assembled using a de novo assembly algorithm within Velvet software (version 1.2.08) (50). Overall, the average number of contiguous sequences (contigs) for all 104 genomes sequenced in this study was 439, which gave rise to an average total assembled genome size of 2,436,856 bp.
Genomes are archived using a gene-by-gene approach for genome alignment and comparison supported by the BLAST algorithm (51). A reference pan-genome was constructed from the clinical isolate genomes (all collected as part of this study) and the reference S. epidermidis RP62A (ATCC 35984) and ATCC 12228 genomes (52). Putative gene function was assigned to genes in the reference pan-genome list using RAST (Rapid Annotations using Subsystem Technology) (53) and the SEED database (54), which were cross-referenced with the S. epidermidis RP62A (ATCC 35984) and ATCC 12228 reference genomes before removing duplicate genes. The BLAST algorithm was used to scan all genomes for gene orthologs at each locus in the reference pan-genome. An ortholog was defined as a reciprocal best hit of the sequence with >70% nucleotide identity over at least 50% of the alignment length. MAFFT software (55) was used to align gene orthologs on a gene-by-gene basis, and these data were concatenated into contiguous sequence for each isolate genome, including gaps. A core genome of 123 genes was defined based on gene presence in all isolates (100%).
Estimating genealogies.
ClonalFrame infers the clonal relationship of bacteria and the chromosomal position of homologous recombination events that disrupt a clonal pattern of inheritance (56). A stringent approach was used to estimate a reduced core genome for construction of a genealogy using ClonalFrame (version 1.2) on concatenated sequences of 104 S. epidermidis genomes with 100,000 iterations, half of which were discarded as burn-in. Substitution mutation and recombination regions were categorized from the output of ClonalFrame. The posterior probability of recombination and substitution at each site is calculated by ClonalFrame, and recombination events were defined with a probability of recombination of more than 75%, reaching 95% at any one site. The trees were visualized and annotated using MEGA6 (57).
Statistical analysis.
Associations among and between the clinical parameters, bacterial phenotypes, clades, and presence/absence of genes were analyzed statistically using the chi-square test, Fisher's exact test, the Cochran-Armitage trend test, or the Kruskal-Wallis test as appropriate. The chi-square test was carried out to test the null hypothesis that the lineages are homogenous in their clinical outcome or resistance phenotypes. Permutation tests were performed to test the null hypothesis that there was no association between lineage and clinical outcome or resistance phenotype. Association between clinical outcome or antimicrobial resistance and lineage in the observed data was summarized by an association score. Statistical analyses were performed using SAS software (version 9.2; SAS, Cary, NC, USA) and SPSS (version 10; IBM, USA), and the level of significance was set at a P value of ≤0.05.
Data availability.
Short reads are available from the NCBI Sequence Read Archive (SRA) associated with BioProject no. PRJNA382527. Assembled genomes are also archived in the publicly accessible Staphylococcal Bacterial Isolate Genome Sequence database (BIGSdb) (https://sheppardlab.com/resources/).
ACKNOWLEDGMENTS
We acknowledge Kathrin Espinoza, AO Clinical Investigation and Documentation, Dübendorf, Switzerland, for statistical support.
This work was funded by AO Trauma as part of the Clinical Priority Program, Bone Infection. S.K.S. is supported by the Medical Research Council, project reference MR/L015080/1.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
All authors have no reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the article have been disclosed.
REFERENCES
- 1.Trampuz A, Zimmerli W. 2005. Prosthetic joint infections: update in diagnosis and treatment. Swiss Med Wkly 135:243–251. [DOI] [PubMed] [Google Scholar]
- 2.Trampuz A, Zimmerli W. 2006. Diagnosis and treatment of infections associated with fracture-fixation devices. Injury 37(Suppl 2):S59–S66. [DOI] [PubMed] [Google Scholar]
- 3.Conlan S, Mijares LA, Program NCS, Becker J, Blakesley RW, Bouffard GG, Brooks S, Coleman H, Gupta J, Gurson N, Park M, Schmidt B, Thomas PJ, Otto M, Kong HH, Murray PR, Segre JA. 2012. Staphylococcus epidermidis pan-genome sequence analysis reveals diversity of skin commensal and hospital infection-associated isolates. Genome Biol 13:R64. doi: 10.1186/gb-2012-13-7-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meric G, Miragaia M, de Been M, Yahara K, Pascoe B, Mageiros L, Mikhail J, Harris LG, Wilkinson TS, Rolo J, Lamble S, Bray JE, Jolley KA, Hanage WP, Bowden R, Maiden MC, Mack D, de Lencastre H, Feil EJ, Corander J, Sheppard SK. 2015. Ecological overlap and horizontal gene transfer in Staphylococcus aureus and Staphylococcus epidermidis. Genome Biol Evol 7:1313–1328. doi: 10.1093/gbe/evv066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miragaia M, Thomas JC, Couto I, Enright MC, de Lencastre H. 2007. Inferring a population structure for Staphylococcus epidermidis from multilocus sequence typing data. J Bacteriol 189:2540–2552. doi: 10.1128/JB.01484-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris LG, Murray S, Pascoe B, Bray J, Meric G, Magerios L, Wilkinson TS, Jeeves R, Rohde H, Schwarz S, de Lencastre H, Miragaia M, Rolo J, Bowden R, Jolley KA, Maiden MC, Mack D, Sheppard SK. 2016. Biofilm morphotypes and population structure among Staphylococcus epidermidis from commensal and clinical samples. PLoS One 11:e0151240. doi: 10.1371/journal.pone.0151240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fey PD, Olson ME. 2010. Current concepts in biofilm formation of Staphylococcus epidermidis. Future Microbiol 5:917–933. doi: 10.2217/fmb.10.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morgenstern M, Post V, Erichsen C, Hungerer S, Buhren V, Militz M, Richards G, Moriarty F. 2016. Biofilm formation increases treatment failure in Staphylococcus epidermidis device-related osteomyelitis of the lower extremity in human patients. J Orthop Res doi: 10.1002/jor.23218. [DOI] [PubMed] [Google Scholar]
- 9.Yao Y, Sturdevant DE, Villaruz A, Xu L, Gao Q, Otto M. 2005. Factors characterizing Staphylococcus epidermidis invasiveness determined by comparative genomics. Infect Immun 73:1856–1860. doi: 10.1128/IAI.73.3.1856-1860.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gu J, Li H, Li M, Vuong C, Otto M, Wen Y, Gao Q. 2005. Bacterial insertion sequence IS256 as a potential molecular marker to discriminate invasive strains from commensal strains of Staphylococcus epidermidis. J Hosp Infect 61:342–348. doi: 10.1016/j.jhin.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 11.Montanaro L, Campoccia D, Pirini V, Ravaioli S, Otto M, Arciola CR. 2007. Antibiotic multiresistance strictly associated with IS256 and ica genes in Staphylococcus epidermidis strains from implant orthopedic infections. J Biomed Mater Res A 83:813–818. doi: 10.1002/jbm.a.31399. [DOI] [PubMed] [Google Scholar]
- 12.Harris LG, Bexfield A, Nigam Y, Rohde H, Ratcliffe NA, Mack D. 2009. Disruption of Staphylococcus epidermidis biofilms by medicinal maggot Lucilia sericata excretions/secretions. Int J Artif Organs 32:555–564. [DOI] [PubMed] [Google Scholar]
- 13.Mack D, Davies AP, Harris LG, Knobloch JK, Rohde H. 2009. Staphylococcus epidermidis biofilms: functional molecules, relation to virulence, and vaccine potential. Top Curr Chem 288:157–182. doi: 10.1007/128_2008_19. [DOI] [PubMed] [Google Scholar]
- 14.Otto M. 2012. Molecular basis of Staphylococcus epidermidis infections. Semin Immunopathol 34:201–214. doi: 10.1007/s00281-011-0296-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas JC, Vargas MR, Miragaia M, Peacock SJ, Archer GL, Enright MC. 2007. Improved multilocus sequence typing scheme for Staphylococcus epidermidis. J Clin Microbiol 45:616–619. doi: 10.1128/JCM.01934-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campoccia D, Montanaro L, Pirini V, Ravaioli S, Arciola CR. 2009. Prevalence of genes for aminoglycoside-modifying enzymes in Staphylococcus epidermidis isolates from orthopedic postsurgical and implant-related infections. J BiomedMater Res A 88:654–663. doi: 10.1002/jbm.a.31754. [DOI] [PubMed] [Google Scholar]
- 17.Otto M. 2009. Staphylococcus epidermidis—the ‘accidental’ pathogen. Nat Rev Microbiol 7:555–567. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spiegl U, Friederichs J, Patzold R, Militz M, Josten C, Buhren V. 2013. Risk factors for failed two-stage procedure after chronic posttraumatic periprosthetic hip infections. Arch Orthop Trauma Surg 133:421–428. doi: 10.1007/s00402-012-1673-6. [DOI] [PubMed] [Google Scholar]
- 19.Uckay I, Pittet D, Vaudaux P, Sax H, Lew D, Waldvogel F. 2009. Foreign body infections due to Staphylococcus epidermidis. Ann Med 41:109–119. doi: 10.1080/07853890802337045. [DOI] [PubMed] [Google Scholar]
- 20.Bowden MG, Chen W, Singvall J, Xu Y, Peacock SJ, Valtulina V, Speziale P, Hook M. 2005. Identification and preliminary characterization of cell-wall-anchored proteins of Staphylococcus epidermidis. Microbiology 151:1453–1464. doi: 10.1099/mic.0.27534-0. [DOI] [PubMed] [Google Scholar]
- 21.Cucarella C, Solano C, Valle J, Amorena B, Lasa I, Penades JR. 2001. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J Bacteriol 183:2888–2896. doi: 10.1128/JB.183.9.2888-2896.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kilgus DJ, Howe DJ, Strang A. 2002. Results of periprosthetic hip and knee infections caused by resistant bacteria. Clin Orthop Relat Res 2002:116–124. doi: 10.1097/00003086-200211000-00021. [DOI] [PubMed] [Google Scholar]
- 23.Salgado CD, Dash S, Cantey JR, Marculescu CE. 2007. Higher risk of failure of methicillin-resistant Staphylococcus aureus prosthetic joint infections. Clin Orthop Relat Res 461:48–53. [DOI] [PubMed] [Google Scholar]
- 24.Leung F, Richards CJ, Garbuz DS, Masri BA, Duncan CP. 2011. Two-stage total hip arthroplasty: how often does it control methicillin-resistant infection? Clin Orthop Relat Res 469:1009–1015. doi: 10.1007/s11999-010-1725-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teterycz D, Ferry T, Lew D, Stern R, Assal M, Hoffmeyer P, Bernard L, Uckay I. 2010. Outcome of orthopedic implant infections due to different staphylococci. Int J Infect Dis 14:e913–8. doi: 10.1016/j.ijid.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 26.Volin SJ, Hinrichs SH, Garvin KL. 2004. Two-stage reimplantation of total joint infections: a comparison of resistant and non-resistant organisms. Clin Orthop Relat Res 2004:94–100. doi: 10.1097/01.blo.0000143559.34143.3d. [DOI] [PubMed] [Google Scholar]
- 27.Anguita-Alonso P, Hanssen AD, Osmon DR, Trampuz A, Steckelberg JM, Patel R. 2005. High rate of aminoglycoside resistance among staphylococci causing prosthetic joint infection. Clin Orthop Relat Res 439:43–47. doi: 10.1097/01.blo.0000182394.39601.9d. [DOI] [PubMed] [Google Scholar]
- 28.Abbassi MS, Bouchami O, Touati A, Achour W, Ben Hassen A. 2008. Clonality and occurrence of genes encoding antibiotic resistance and biofilm in methicillin-resistant Staphylococcus epidermidis strains isolated from catheters and bacteremia in neutropenic patients. Curr Microbiol 57:442–448. doi: 10.1007/s00284-008-9227-4. [DOI] [PubMed] [Google Scholar]
- 29.Cherifi S, Byl B, Deplano A, Nonhoff C, Denis O, Hallin M. 2013. Comparative epidemiology of Staphylococcus epidermidis isolates from patients with catheter-related bacteremia and from healthy volunteers. J Clin Microbiol 51:1541–1547. doi: 10.1128/JCM.03378-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wassenaar TM, Ussery D, Nielsen LN, Ingmer H. 2015. Review and phylogenetic analysis of qac genes that reduce susceptibility to quaternary ammonium compounds in Staphylococcus species. Eur J Microbiol Immunol 5:44–61. doi: 10.1556/EuJMI-D-14-00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prag G, Falk-Brynhildsen K, Jacobsson S, Hellmark B, Unemo M, Soderquist B. 2014. Decreased susceptibility to chlorhexidine and prevalence of disinfectant resistance genes among clinical isolates of Staphylococcus epidermidis. APMIS 122:961–967. doi: 10.1111/apm.12239. [DOI] [PubMed] [Google Scholar]
- 32.Taheri N, Ardebili A, Amouzandeh-Nobaveh A, Ghaznavi-Rad E. 2016. Frequency of antiseptic resistance among Staphylococcus aureus and coagulase-negative staphylococci isolated from a university hospital in central Iran. Oman Med J 31:426–432. doi: 10.5001/omj.2016.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Culebras E, Martinez JL. 1999. Aminoglycoside resistance mediated by the bifunctional enzyme 6′-N-aminoglycoside acetyltransferase-2″O-aminoglycoside phosphotransferase. Front Biosci 4:D1–D8. [DOI] [PubMed] [Google Scholar]
- 34.Thomas WD Jr, Archer GL. 1989. Mobility of gentamicin resistance genes from staphylococci isolated in the United States: identification of Tn4031, a gentamicin resistance transposon from Staphylococcus epidermidis. Antimicrob Agents Chemother 33:1335–1341. doi: 10.1128/AAC.33.8.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozitskaya S, Cho SH, Dietrich K, Marre R, Naber K, Ziebuhr W. 2004. The bacterial insertion sequence element IS256 occurs preferentially in nosocomial Staphylococcus epidermidis isolates: association with biofilm formation and resistance to aminoglycosides. Infect Immun 72:1210–1215. doi: 10.1128/IAI.72.2.1210-1215.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ziebuhr W, Krimmer V, Rachid S, Lossner I, Gotz F, Hacker J. 1999. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol Microbiol 32:345–356. doi: 10.1046/j.1365-2958.1999.01353.x. [DOI] [PubMed] [Google Scholar]
- 37.Galdbart JO, Morvan A, El Solh N. 2000. Phenotypic and molecular typing of nosocomial methicillin-resistant Staphylococcus aureus strains susceptible to gentamicin isolated in france from 1995 to 1997. J Clin Microbiol 38:185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bogut A, Niedzwiadek J, Koziol-Montewka M, Strzelec-Nowak D, Blacha J, Mazurkiewicz T, Marczynski W, Plewik D. 2014. Characterization of Staphylococcus epidermidis and Staphyloccocus warneri small-colony variants associated with prosthetic-joint infections. J Med Microbiol 63:176–185. doi: 10.1099/jmm.0.066068-0. [DOI] [PubMed] [Google Scholar]
- 39.Hirschhausen N, Block D, Bianconi I, Bragonzi A, Birtel J, Lee JC, Dubbers A, Kuster P, Kahl J, Peters G, Kahl BC. 2013. Extended Staphylococcus aureus persistence in cystic fibrosis is associated with bacterial adaptation. Int J Med Microbiol 303:685–692. doi: 10.1016/j.ijmm.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 40.Proctor RA, Kriegeskorte A, Kahl BC, Becker K, Loffler B, Peters G. 2014. Staphylococcus aureus small colony variants (SCVs): a road map for the metabolic pathways involved in persistent infections. Front Cell Infect Microbiol 4:99. doi: 10.3389/fcimb.2014.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sendi P, Proctor RA. 2009. Staphylococcus aureus as an intracellular pathogen: the role of small colony variants. Trends Microbiol 17:54–58. doi: 10.1016/j.tim.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 42.Singh R, Ray P, Das A, Sharma M. 2009. Role of persisters and small-colony variants in antibiotic resistance of planktonic and biofilm-associated Staphylococcus aureus: an in vitro study. J Med Microbiol 58:1067–1073. doi: 10.1099/jmm.0.009720-0. [DOI] [PubMed] [Google Scholar]
- 43.Tuchscherr L, Kreis CA, Hoerr V, Flint L, Hachmeister M, Geraci J, Bremer-Streck S, Kiehntopf M, Medina E, Kribus M, Raschke M, Pletz M, Peters G, Loffler B. 2016. Staphylococcus aureus develops increased resistance to antibiotics by forming dynamic small colony variants during chronic osteomyelitis. J Antimicrob Chemother 71:438–448. doi: 10.1093/jac/dkv371. [DOI] [PubMed] [Google Scholar]
- 44.Tuchscherr L, Loffler B. 2016. Staphylococcus aureus dynamically adapts global regulators and virulence factor expression in the course from acute to chronic infection. Curr Genet 62:15–17. doi: 10.1007/s00294-015-0503-0. [DOI] [PubMed] [Google Scholar]
- 45.von Eiff C, McNamara P, Becker K, Bates D, Lei XH, Ziman M, Bochner BR, Peters G, Proctor RA. 2006. Phenotype microarray profiling of Staphylococcus aureus menD and hemB mutants with the small-colony-variant phenotype. J Bacteriol 188:687–693. doi: 10.1128/JB.188.2.687-693.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perez K, Patel R. 2017. Staphylococcus epidermidis small-colony variants are induced by low pH and their frequency reduced by lysosomal alkalinization. J Infect Dis 215:488–490. doi: 10.1093/infdis/jiw503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tande AJ, Osmon DR, Greenwood-Quaintance KE, Mabry TM, Hanssen AD, Patel R. 2014. Clinical characteristics and outcomes of prosthetic joint infection caused by small colony variant staphylococci. mBio 5:e01910-14. doi: 10.1128/mBio.01910-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Post V, Wahl P, Uckay I, Ochsner P, Zimmerli W, Corvec S, Loiez C, Richards RG, Moriarty TF. 2014. Phenotypic and genotypic characterisation of Staphylococcus aureus causing musculoskeletal infections. Int J Med Microbiol 304:565–576. doi: 10.1016/j.ijmm.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 49.Stepanovic S, Vukovic D, Hola V, Di Bonaventura G, Djukic S, Cirkovic I, Ruzicka F. 2007. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS 115:891–899. doi: 10.1111/j.1600-0463.2007.apm_630.x. [DOI] [PubMed] [Google Scholar]
- 50.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sheppard SK, Jolley KA, Maiden MC. 2012. A gene-by-gene approach to bacterial population genomics: whole genome MLST of Campylobacter. Genes (Basel) 3:261–277. doi: 10.3390/genes3020261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meric G, Yahara K, Mageiros L, Pascoe B, Maiden MC, Jolley KA, Sheppard SK. 2014. A reference pan-genome approach to comparative bacterial genomics: identification of novel epidemiological markers in pathogenic Campylobacter. PLoS One 9:e92798. doi: 10.1371/journal.pone.0092798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res 42:D206–D14. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Didelot X, Falush D. 2007. Inference of bacterial microevolution using multilocus sequence data. Genetics 175:1251–1266. doi: 10.1534/genetics.106.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Short reads are available from the NCBI Sequence Read Archive (SRA) associated with BioProject no. PRJNA382527. Assembled genomes are also archived in the publicly accessible Staphylococcal Bacterial Isolate Genome Sequence database (BIGSdb) (https://sheppardlab.com/resources/).