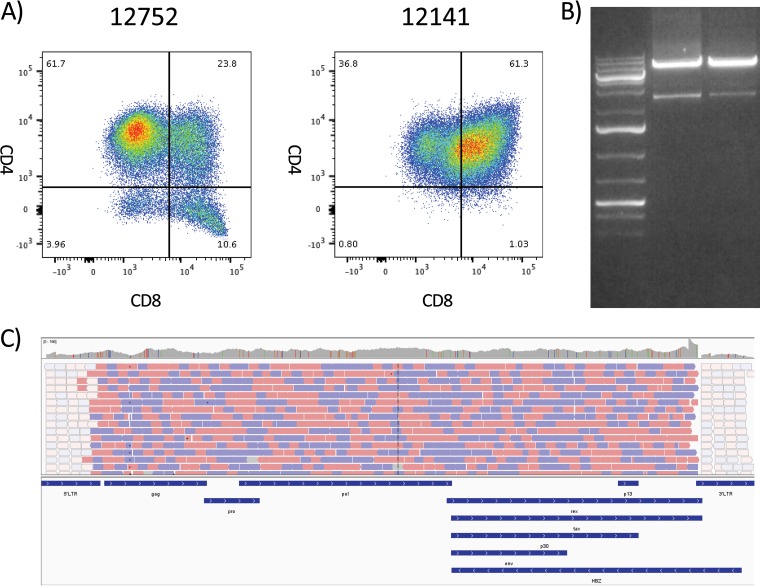
FIG 1.
PCR amplification and sequencing of the whole STLV-1 genome from STLV-positive T cell lines. Genomic DNA was purified from T cell lines grown out of STLV-positive baboon PBMC samples. (A) The cell line phenotype was determined by flow cytometry, gating on the lymphocyte population, and standard CD3, CD4, and CD8 T cell staining. The cell line of animal 12752 was predominantly made up of CD4+ CD8− T cells, whereas the cell line of animal 12141 displayed a large degree of double-positive CD4+ CD8+ T cells. The numbers in each quadrant represent the percentage of live CD3+ lymphocytes in that quadrant. (B) PCR was performed with an ultra-high-fidelity DNA polymerase with 25 rounds of amplification to minimize PCR error. PCR products were run on an agarose gel, and the 8.5-kb band was excised and purified for NGS. (C) Next-generation sequencing was performed on a HiSeq 2500 desktop sequencer with the assistance of the University of Miami’s John P. Hussman Institute for Human Genomics sequencing core. Sequencing provided almost complete genome coverage, with only a small portion of the LTR being excluded. Animal 12752 provided 21.2 million quality reads, whereas animal 12141 provided 23.5 million quality reads.