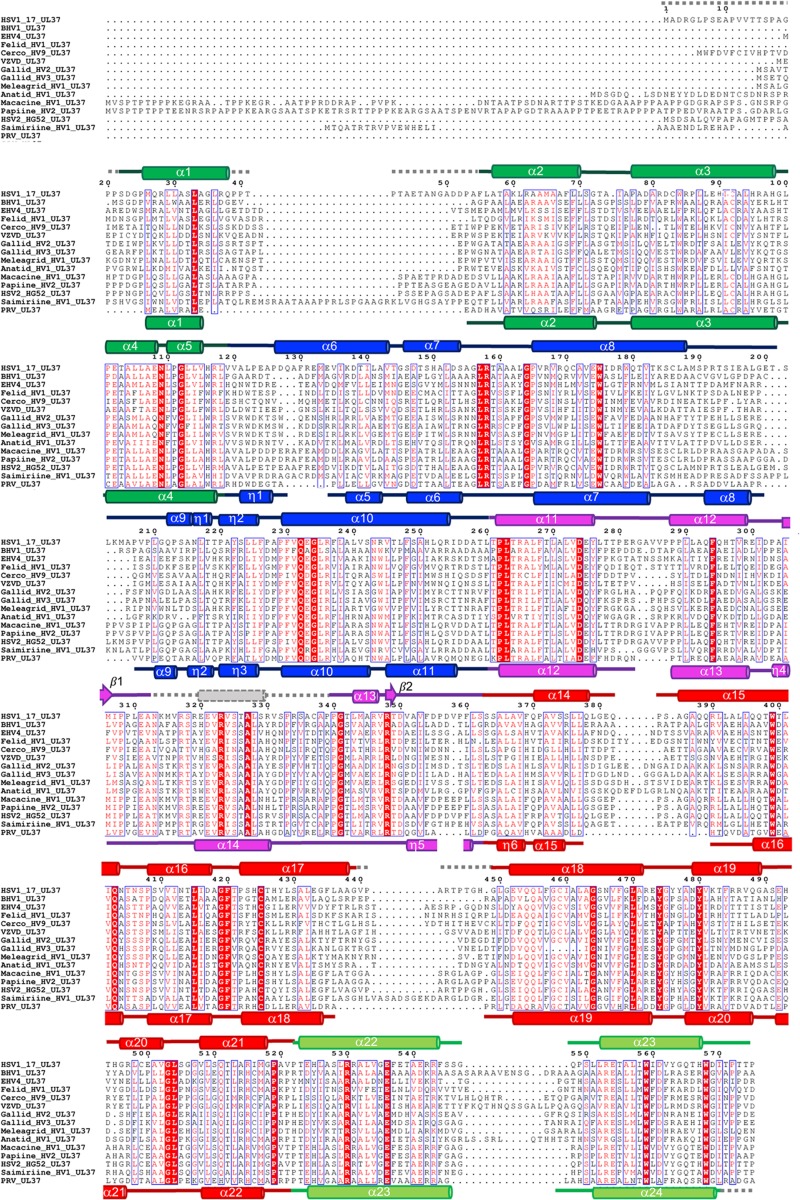
FIG 2.
Multiple-sequence alignment of UL37N from 15 alphaherpesviruses. Sequence alignment was generated using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). Only the alignment of residues corresponding to residues 1 to 575 of HSV-1 UL37 is shown. Identical residues are shown in white letters on a red background. Similar residues are shown as red letters boxed in blue. Secondary structure derived from the crystal structures of HSV-1 and PRV UL37 homologs is shown above and below the alignment, respectively. Helices are represented by cylinders and beta strands by arrows. Secondary structure elements are labeled and colored by domain using the same color scheme as in Fig. 1. Unresolved residues are represented using gray dashed lines. HSV-1 and PRV UL37N share 29.7% identity and 55.9% similarity. Sequences from the following viruses were used (GenBank Gene ID numbers are given in parentheses): HSV-1_17, herpes simplex virus 1 strain 17 (2703358); BHV1, bovine herpesvirus 1 (1487388); EHV4, equine herpesvirus 4 (1487587); Felid_HV1, felid herpesvirus 1 (8658550); Cerco_HV9, cercopithecine herpesvirus 9 (920525); VZVD, varicella-zoster virus serotype D (1487686); Gallid_HV2, gallid herpesvirus 2 (4811511); Gallid_HV3, gallid herpesvirus 3 (911942); Meleagrid_HV1, meleagrid herpesvirus 1 (918517); Anatid_HV1, anatid herpesvirus 1 (8223349); Macacine_HV1, macacine herpesvirus 1 (1487432); Papiine_HV2, papiine herpesvirus 2 (3850220); HSV2_HG52, herpes simplex virus 2 strain HG52 (1487323); Saimiriine_HV1, saimiriine herpesvirus 1 (9829319); PRV, suid herpesvirus strain Kaplan (2952491).