FIG 2.
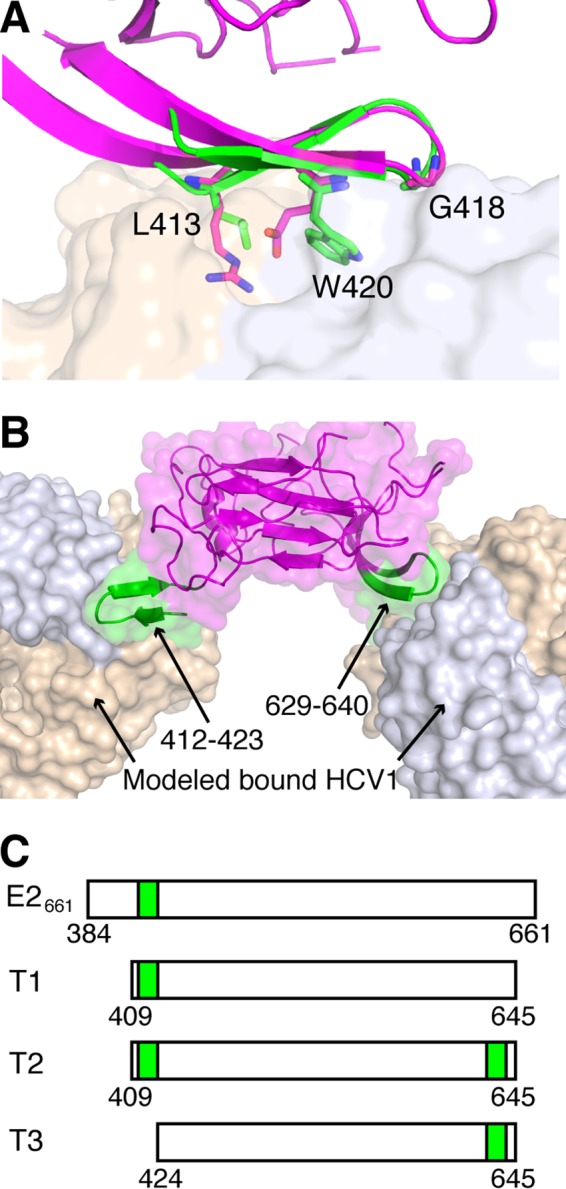
Design of an E2-based epitope I bivalent immunogen. (A) Structure of E2 core (PDB code 4MWF; magenta), aligned to HCV1-bound epitope I (PDB code 4DGY; green) using residues 629 to 640. HCV1 MAb (tan and light blue surface for heavy and light chains, respectively) is shown for reference. Epitope residues L413, G418, and W420, as well as corresponding E2 core residues R630, G635, and E637, are shown as sticks, and epitope positions are labeled. The backbone root mean square distance between epitope I and matching E2 core residues (aa 629 to 640) is 0.8 Å. (B) Model of HCV1 MAb bound to the native epitope I site on E2 (aa 412 to 423) as well as engineered epitope I site at aa 629 to 640, with both sites colored green, and E2 core and MAb colored as in panel A. (C) Tested E2 constructs E2661, T1 (truncated native), T2 (truncated bivalent), and T3 (truncated engineered site only), with E2 epitope I sequence locations represented as green boxes.
