FIG 3.
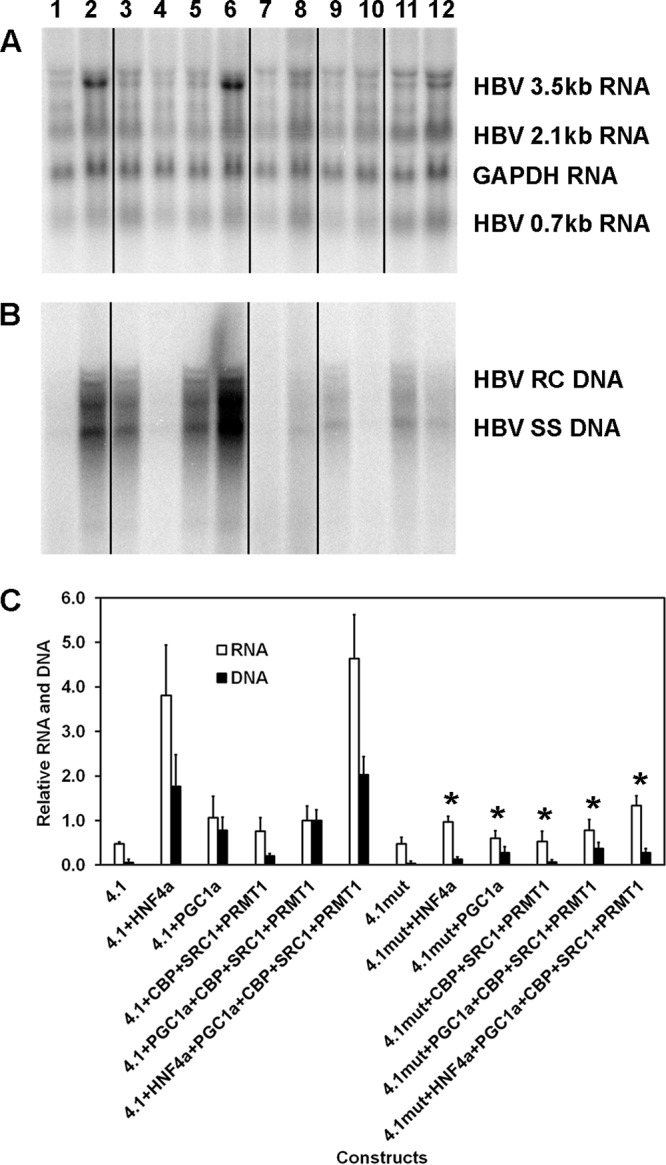
Role of the proximal nuclear receptor binding site in transcriptional coactivator-mediated activation of HBV biosynthesis in the human embryonic kidney cell line HEK293T. (A) RNA (Northern) filter hybridization analysis of HBV transcripts. The 3.9-kb transcript observed above the HBV 3.5-kb RNA probably represents the previously reported HBV long xRNA that initiates from the X promoter region (55). The GAPDH transcript was used as an internal control for RNA loading per lane. The black lines indicate noncontiguous lanes from a single filter hybridization analysis. (B) DNA (Southern) filter hybridization analysis of HBV replication intermediates. Cells were transfected with the HBV DNA (4.1-kbp) or HBVHNF4mut DNA (4.1-kbp) constructs (lanes 1 to 6 and 7 to 12, respectively) plus the HNF4α (lanes 2 and 8), PGC1α (lanes 3, 5, 6, 9, 11, and 12), CBP, SRC1, and PRMT1 (lanes 4 to 6 and 10 to 12) expression vectors. The black lines indicate noncontiguous lanes from a single filter hybridization analysis. (C) Quantitative analysis of the HBV 3.5-kb RNA and HBV DNA replication intermediates. The levels of the HBV 3.5-kb RNA and total HBV DNA replication intermediates are reported relative to the value for HBV DNA (4.1-kbp) construct in the presence of the expression of the four coactivators (lane 5). Mean RNA and DNA levels plus standard deviations from five independent analyses are shown. Levels of the transcripts (lanes 8 and 12) and replication intermediates (lanes 8 to 12) in HBVHNF4mut DNA (4.1-kbp) construct-transfected cells (lanes 7 to 12) that are statistically significantly lower than the levels in HBV DNA (4.1-kbp) construct-transfected cells expressing the same transcription factors and/or coactivators (lanes 1 to 6), as determined by Student's t test (P < 0.05), are indicated with an asterisk.
