FIG 5.
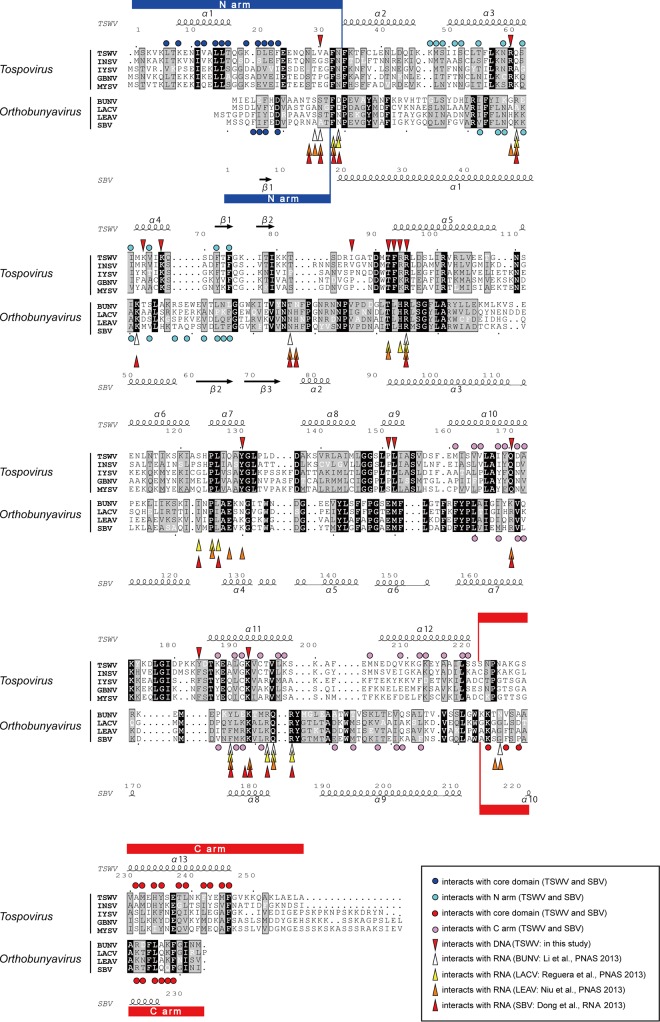
Amino acid sequence alignment of N proteins among tospoviruses and orthobunyaviruses. Key residues for nucleic acid binding are marked with downward arrowheads (red, TSWV) (the residues interact with DNA molecules) and upward arrowheads (white, Bunyamwera virus (BUNV); yellow, LACV; orange, Leanyer virus (LEAV); red, SBV). Key residues in the N arm for the interactions with the core domain are marked with dark blue circles, and key residues in the core domain for the interactions with the N arm are marked with light blue circles (TSWV and SBV). Key residues in the C arm for the interactions with the core domain are marked with red circles, and key residues in the core domain for the interactions with the C arm are marked with pink circles (TSWV and SBV). For each genus, strictly conserved residues are depicted in white characters on a black background, and highly conserved residues are depicted as black characters on a gray background. At the top and bottom of the sequences, a schematic representation of the secondary structure elements of TSWV NP and SBV NP are shown, and every 10 residues of TSWV NP and SBV NP are indicated with a dot. Basically, the alignment was generated by using ClustalW (57). The α-helix is depicted by a coil, and the β-strand is depicted by an arrow. The figure was generated by using ESPript (58) and modified manually based on the structural information. The N arm is underscored in blue, and the C arm is underscored in red. GenBank database accession numbers are as follows: AB889601 for TSWV NP, AFH88369 for Impatiens necrotic spot virus (INSV) NP, ACA09432 for Iris yellow spot virus (IYSV) NP, NP_619701 for Groundnut bud necrosis virus (GBNV) NP, ABY60853 for Melon yellow spot virus (MYSV) NP, NP_047213 for BUNV NP, P04873 for LACV NP, AEA02984 for LEAV NP, and CCF55031 for SBV NP (24–27).