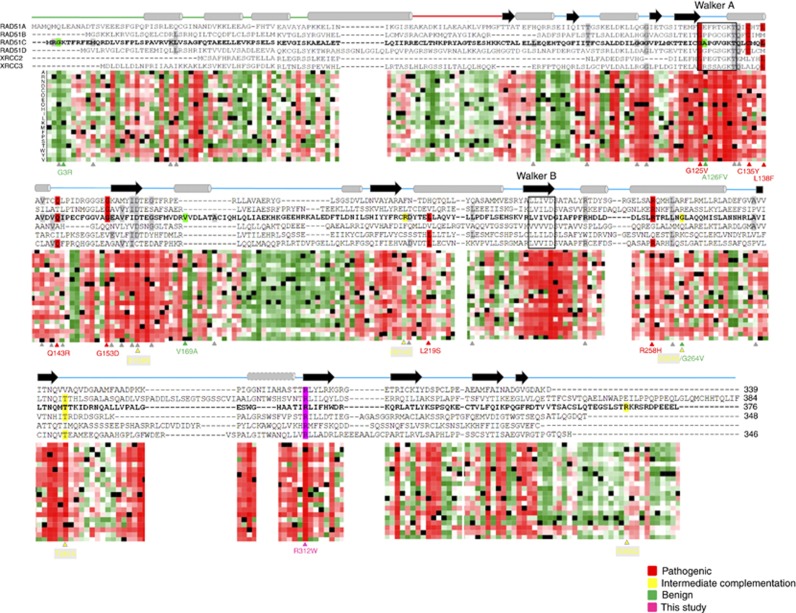
Figure 5.
In silico studies. Sequence alignment of human RAD51C and its paralogues RAD51A, RAD51B, RAD51D, XRCC2 and XRCC3. Secondary structure obtained from P.furiosus (Pf) RAD51 and described by Miller et al (2004) is shown schematically above the sequences with cylinders for α-helices and arrows for β-strands. The helix showed with a dashed line is predicted in the Rad51 paralogues using PSI-PRED, but is absent in the P. furiosus Rad51 crystal structure. The Walker A and Walker B ATP binding motifs are indicated by rectangles. Horizontal line linking α-helixes and β-strands correspond to the N-terminal domain (green), linker region (red) and C-terminal domain (blue). A heat map that shows the tolerance to independent amino acid (aa) substitutions (y-axis) for each position of the RAD51C protein (x-axis) generated using PredictProtein tool (Yachdav et al, 2014) is shown below the sequence alignment. Red indicates strong signal for a deleterious effect (score>50); white indicates a small effect; green indicates a strong signal for neutral effect/no effect (score<−50); and black represents the corresponding wild-type residue. RAD51C missense substitutions functionally characterised described in the literature and listed in Table 1 are specified below the heatmap and shown in different colours: Residues highlighted in red, green and yellow correspond to pathogenic, benign and undetermined/hypomorphic missense variants, respectively. Variants highlighted in grey correspond to the functionally uncharacterised changes detailed in Table 1. The novel RAD51C p.Arg312Trp pathogenic variant described in this study is highlighted in purple. A full colour version of this figure is available at the British Journal of Cancer journal online.