Abstract
Avermectins, a group of polyketide natural products, are widely used as anthelmintics in agriculture. Metabolic engineering and combinatorial biosynthesis were extensively employed to improve Avermectins production and create novel Avermectin derivatives, including Ivermectin and Doramectin. It is labor intensive and time cost to genetically manipulate Avermectins producer Streptomyces avermitilis in vivo. Cloning and heterologous expression of Avermectins biosynthetic gene cluster will make it possible to tailor the cluster in vitro. We constructed a Bacterial Artificial Chromosome (BAC) library of S. avermitilis ATCC 31267 with inserted DNA fragments ranged from 100 to 130 Kb. Five recombinant BAC clones which carried the Avermectins biosynthetic gene cluster ave (81 Kb in size) were screened out from the library. Then, ave was hetero-expressed in S. lividans. Three Avermectin components, A2a, B1a and A1a were detected from the cell extracts of recombinant strains. It will facilitate the development of Avermectin derivatives by polyketide synthase domain swapping and provide functional element for Avermectins synthetic biology study.
Keywords: Streptomyces, Avermectin, BAC library, Heterologous expression
1. Introduction
Streptomyces, a representative genus of filamentous Gram-positive Actinobacteria, are known for the ability to produce secondary metabolites [1]. Bioactive natural products play an important role in medicine and agriculture. It was widely used as antibacterials (e.g Daptomycin) [2], antifungals (e.g Amphotericin B) [3], anthelmintics (e.g Avermectin, chemical structure was shown in Fig. 1) [4], herbicides (e.g Phosphinothricin) [5], immunosuppressants (e.g FK-506) [6], and anticancer agents (e.g Doxorubicin) [7].
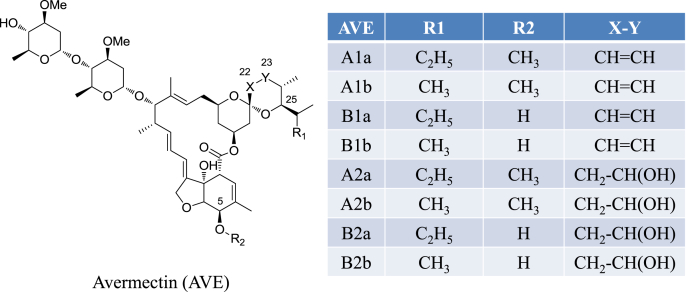
Fig. 1.
Chemical structures of Avermectins.
Generally, microbial natural products biosynthesis related genes locate in linked loci. Therefore, it was defined as biosynthetic gene cluster. The gene cluster size ranges from tens of kilo base-pairs (Kb) to more than 100 Kb. To certain polyketides (PKs) and non-ribosomal peptides (NRPs) natural products, its biosynthetic gene cluster size is really large. For example, the biosynthetic gene cluster of Daptomycin is 128 Kb [8], Amphotericin's is 113 Kb [9], and Avermectin's is 81 Kb (ave, as shown in Fig. 2) [10]. So, large size DNA fragment manipulation was frequently employed in gene cluster tailoring, metabolic engineering, combinatorial biosynthesis and synthetic biology.
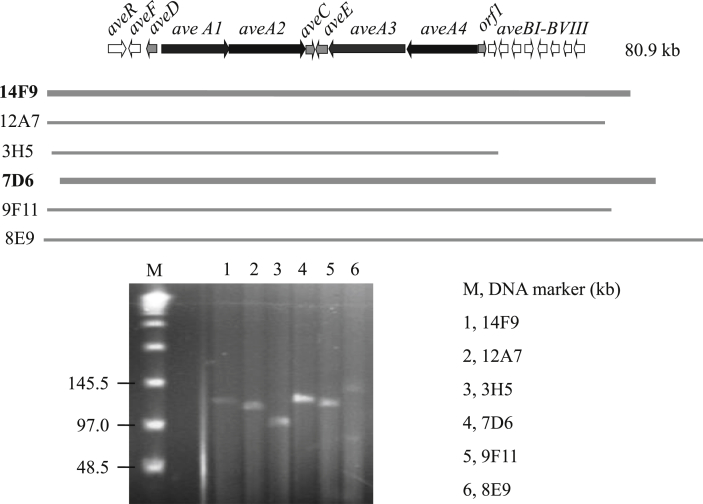
Fig. 2.
BAC recombinants covered Avermectins biosynthetic gene cluster. 14F9, 12A7, 3H5, 7D6, 9F11 and 8E9 meant the number of BAC recombinants.
In the development of Avermectin derivatives, the ave cluster was always tailored in vivo. To make S. avermitilis produce Doramectins, the Avermectins polyketide synthase (PKS) loading module Acyl Transferase (AT) domain was substituted by Phoslactomycin B (or Ansatrienin) AT domain [11], [12]. To produce Ivermectins, domain swapping was carried out in Avermectins PKS module 2 [13], [14]. To produce Milbemycins, Avermectins PKS AveA1 and AveA3 were replaced by Milbemycins PKS MilA1 and MilA3, respectively [15]. It is labor intensive and time cost to get a double cross-over mutant of S. avermitilis. Additionally, missed target cross-over happened frequently since the polyketide synthase DNA sequences are highly similar. To clone the ave cluster and tailor it in vitro will circumvent these obstacles effectively.
Shuttle vectors with high loading capacity are desired approaches for Streptomyces large size DNA fragments clone. Integration vector pSET152 derived from ФC31 phage is one of the most widely used shuttle vectors from Escherichia coli to Streptomyces [16]. Recombinant plasmids derived from pSET152 could be transferred into Streptomyces by conjugation with the help of helping plasmid pUZ8002 [17]. Then, the recombinant plasmids could integrate into the attB site of Streptomyces chromosome by an int/attP integration system [18]. Based on the ground that E. coli replication system of pSET152 is coming from pUC18, its loading capacity is limited. Bacterial artificial chromosome (BAC) vector is a kind of high loading capacity vectors derived from E. coli F plasmid. The inserted DNA fragments size could range from 0 to more than 300 Kb [19]. It maintains steadily at low copy numbers per cell depending on the replication and partition system of F plasmid.
Combination of the removable and integratable ability of pSET152 with the high loading capacity of BAC vector would facilitate the manipulation of Streptomyces large size DNA fragments. A BAC library was constructed for the Avermectins producing strain S. avermitilis ATCC 31267 in this study. The biosynthetic gene cluster of Avermectin was cloned and heterologously expressed successfully.
2. Materials and methods
2.1. DNA manipulations
Plasmid pHZAUBAC1 was digested with BamHI to release BAC vector pIndigoBAC536-S [20]. It was further dephosphorylated with CIAP to prevent self-ligation. S. avermitilis ATCC 31267 spores were inoculated into TSBY (tryptone soya broth 3%, yeast extract 0.5%) medium for seeds preparation. The seed cells were transferred into modified YEME medium (tryptone 0.5%, glucose 1%, malt extract 0.3%, yeast extract 0.3%, 10.3% sucrose, 0.5% glycine) and cultured at 28 °C for 2 days. Mycelia were harvested and treated with 1 mg/mL lysozyme at 30 °C for 50 min, then embedded into low melting point Agarose to construct plugs. It was further treated with 1 mg/mL protease K at 55 °C for 48 h. The genomic DNA size and concentration was evaluated by Pulsed Field Gel Electrophoresis (PFGE). S. avermitilis ATCC 31267 genomic DNA is phosphorothioated, in which the non-bridging oxygen of the ribose phosphate skeleton is substituted by a sulfur [21], [22]. It is challenging to prepare high quality large fragment DNA since it is prone to degradated during the electrophoresis process [23]. A modified TBE buffer (Tris-boric acid 45 mM, EDTA 1 mM) which contains extra 50 μM sulfocarbamide was employed in all of the electrophoresis to protect DNA from degradation. High quality DNA plugs were partial digested with Sau3AI and 120 ± 20 Kb DNA fragments were extracted for library construction [24]. Foreign DNA was ligated with pIndigoBAC536-S vector and transformed into E. coli DH10B competent cells by electroporation as described by our previous work [25].
BAC clones which covered the complete Avermectins biosynthetic gene cluster were screened out by PCR with four primer pairs, aveD-F/R (GGTCAGCACGACATAGC CGA/AAGCTGAGAGGCATCACGGG), aveA1-F/R (CGGGCATGGGAAGGGAA CTT/AACGTCCACCGGGATCATGC), aveA4-F/R (GCACTGATGCGATTTCTGT CAGCCA/CGTGTCGAGCGTGTACCAGTACTC) and Yerd1-F/R (CGGCAAGC CGACCGGCTTCAAACTCTGCCC/TCACACGGTGAAACGGTCGG GGTCGGCGG), respectively. The chloramghenicol resistant gene cat of BAC 14F9 was replaced by a 4.4 Kb int-attP-oriT-acc(3)IV DNA fragment amplified with pSET152 as template and ave-BAC-F (AAGAAACAGATAAATCTTCTGCGTGTGCGGGGGTT GTGGAGGGGCACACGGCGAGTTGGTAGCTCTTGATCCGGCAAACAAACCACC) and ave-BAC-R (GCCTTTTTAAAGACCGTAAAGAAAAATAAGCACAAGT TTTATCCGGCCTTATCGAAGCTGAAAGCACGAGATTCTTCGCCCTGCG) as primers (underlines meant homology arms). PCR targeting was carried out to give BAC 14F9S. BAC 14F9S was conjugated into Streptomyces host cells according to a standard protocol [18].
2.2. Preparation of Avermectins
The sporulation medium for Streptomyces was MS (Soy flour 2%, D-mannitol 2%, agar 2%, pH 7.2), and the liquid medium was YEME (tryptone 0.5%, glucose 1%, malt extract 0.3%, yeast extract 0.3%, 10.3% sucrose). If necessary, apramycin was added with a concentration of 50 μg/mL for MS or YEME medium. S. avermitilis and S. lividans 1326/14F9S were spread on MS plates and incubated at 28 °C for 3 days. Single colony was picked up and inoculated into YEME medium and cultured at 28 °C, 200 rpm for 3 days. 5 mL of the mixture was transferred to 250 mL Erlenmeyer flask containing 50 mL production medium (50.0 g soluble starch, 10.0 g corn flour, 10.0 g yeast extract, 10.0 g soybean flour, 5.0 mg CoCl2·6H2O, 2.0 g CaCO3, per liter, pH 7.2) and cultured at 28 °C, 200 rpm for 7 days [26].
Mycelia were collected by centrifugation and re-suspended by methanol at a ratio of 1.0 g mycelia with 1 mL methanol. The system was homogenized by ultra-sonication at 40 KHz for 30 min and precipitations were eliminated by centrifugation at 8000 g for 20 min. Supernatants were dried out by rotary evaporator and re-suspended by 0.5 mL HPLC grade methanol. The samples were centrifuged at 12,000 g for 30 min and filtered upon subjected to HPLC or LC-MS analysis.
2.3. HPLC and LC-MS analysis of Avermectins
All HPLC analyses were carried out on a Shimadzu (Kyoto, Japan) HPLC instrument equipped with a degasser (DGU-20A3), an auto-sampler (SIL-20A), a column oven (CTO-20A) and two pumps (LC-20AT). Avermectins were tested by monitoring absorption wave length at 245 nm by a PDA detector (SPD-M20A) with commercial Avermectin B1a standards (Sigma-Aldrich) as control. Chromatography was carried out on a Dikma C18 column (5 μm, 4.6 × 25 cm) with a mobile phase was methanol: water = 88: 12, flow rate was 1.0 mL/min, and running time was 30 min. Mass spectrometric analysis was conducted on a Thermo Scientific LTQ - Orbitrap high - resolution mass spectrometer with a positive ion mode scanning from m/z 200 to 2000 as described previously [27].
3. Results
3.1. Construction of the BAC library of S. avermitilis ATCC 31267
Vector pIndigoBAC536-S (7037 bp) is an upgraded BAC vector in which the two NotI restriction sites were replaced by homing endonuclease I-SceI sites and the λ phage cosN and P1 phage loxP sites were removed [17]. To facilitate BAC vectors preparation process, pIndigoBAC536-S was loaded into a high copy number vector pGEM-4Z to generate pHZAUBAC1. Intact pIndigoBAC536-S vectors could be released from pHZAUBAC1 when it was digested with HindIII, BamHI, or EcoRI as described in our previous work [20]. Genome library of Avermectin producing strain S. avermitilis ATCC 31267 was constructed based on pIndigoBAC536-S vector with BamHI as multiple cloning site.
Thirty BAC clones were picked up randomly and the BAC recombinant plasmids were linearized by I-SceI upon PFGE electrophoresis analysis. The inserted DNA fragments ranged from 100 to 130 Kb in size. Five recombinant BAC plasmids, 14F9, 12A7, 7D6, 9F11 and 8E9, covering the complete biosynthetic gene cluster of Avermectins were screened out by PCR from 470 BAC clones. BAC contigs was built up by end sequencing and DNA alignment (as shown in Fig. 2).
3.2. Heterologous expression of Avermectins biosynthetic gene cluster ave in S. lividans 1326
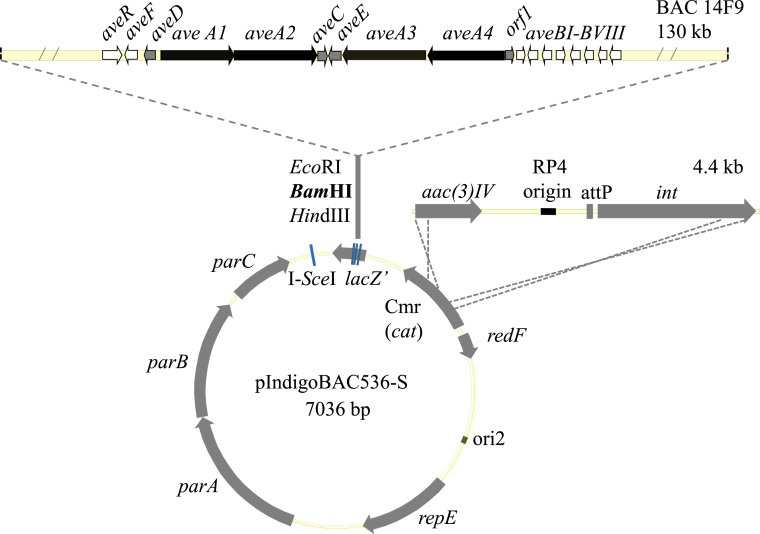
For the heterologous expression of ave, 14F9 was picked out and the chloramghenicol resistant gene cat was replaced by a 4.4 Kb int-attP-oriT-acc(3)IV DNA fragment from pSET152 by PCR targeting. The modified BAC recombinant plasmid 14F9 was defined as 14F9S (map as shown in Fig. 3). To it, int/attP is the integration system, oriT is the conjugation initiation site and acc(3)IV is an Apramycin resistance encoding gene [16].
Fig. 3.
Physical map of BAC recombinant 14F9S derived from pIndigoBAC536-S.
Benefit from the strategic design that there is a conjugation initiation site oriT planted in 14F9S, conjugational transfer could be employed to circumvent protoplast transformation, which is a kind of challenging experiment for large size plasmids. 14F9S was transferred into E. coli ETU12567, which harbors a helping plasmid pUZ8002 [17]. Then, it was conjugated into a wide range of hosts, including S. albus J-1074, S. lividans 1326 and S. coelicolor M145. The integratable characters of 14F9S blocked the possibility of detecting free plasmids from Streptomyces cells directly. Identification of the biosynthetic product of Avermectins would be a realizable strategy to check the heterologous expression system.
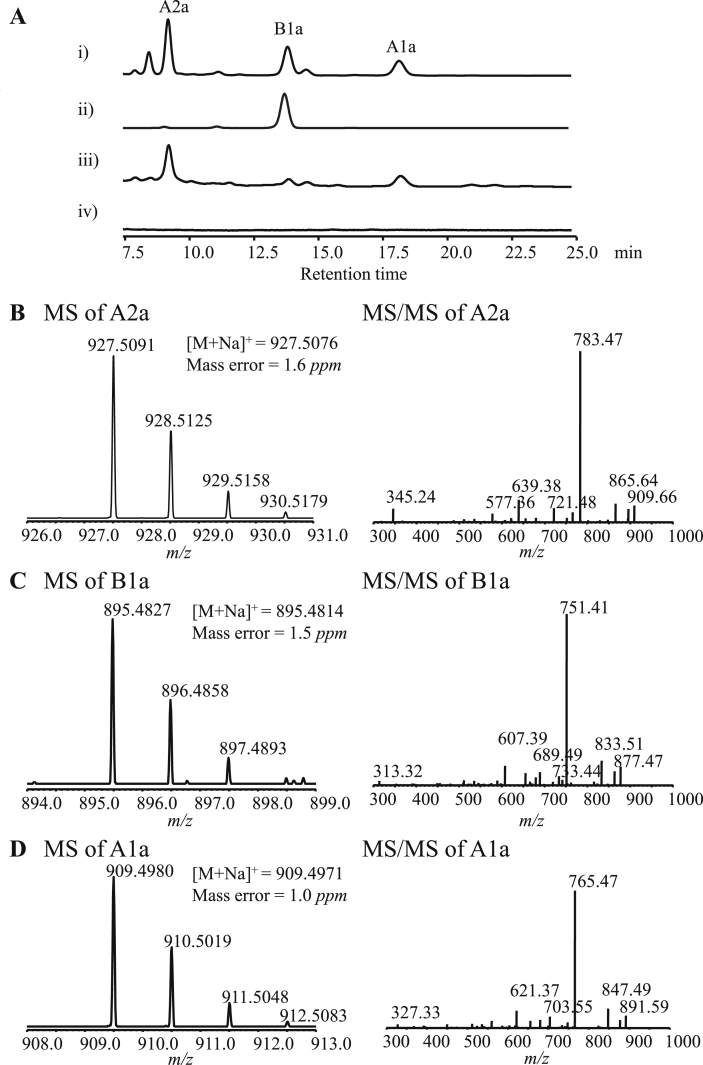
Cell extractions of S. lividans 1326 harboring recombinant BAC plasmid 14F9S were analyzed by HPLC-MS. HPLC results showed that there were three components produced by S. lividans 1326/14F9S accorded well with Avermectins A2a, B1a and A1a produced by Avermectins wild type producers S. avermitilis ATCC 31267. The produced B1a component showed exactly identical retention time with that of B1a standards (Fig. 4A). Furthermore, high resolution mass spectra of the three components accord well with the theoretical exact mass of Avermectins A2a, B1a and A1a, respectively (Fig. 4B, C and 4D). Fragmentation patterns of B1a had been well examined since it is the most active and most widely used component. The observed daughter fragments of B1a were identical to the documented ones. It indicated that S. lividans 1326/14F9S produced three Avermectin components A2a, B1a and A1a successfully. The Avermectins production of recombinant strain S. lividans 1326/14F9S was close to that of Avermectins wild producer S. avermitilis ATCC 31267. A2a was the main one among all detected components (Fig. 4A). To the other five components of Avemectins, it was not detected due to the low titer here. Not every host was qualified cell for ave heterologous expression. Avemectins was not detected from the cell extracts of S. albus or S. coelicolor. Maybe, it is because of the distinct physiological background of certain host cells.
Fig. 4.
HPLC-MS analysis of Avermectins. A, HPLC profile of Avermectins. i) S. avermitilis ATCC 31267, ii) Avermectin B1a standards, iii) S. lividans 1326/14F9S, iv) S. lividans 1326. B, mass spectra of Avermectin component A2a. C, mass spectra of Avermectin component B1a. D, mass spectra of Avermectin component A1a.
4. Discussion
Avermectins have attracted constant attentions since it was discovered in the middle of 1970′ based on the fact that it is an agricultural and medical useful compound. The discoverers of Avermectins shared Nobel Prize in Physiology or Medicine with that of Artemisinin in 2015. Massive efforts had carried out to improve Avermectins production [28], [29], [30], and create Avermectins derivatives, including Ivermectins [13], [14], Doramectins [11], [12], and Milbemycins [15]. It is labor intensive and time cost in metabolic engineering Avermectins native producers and tailoring Avermectins biosynthetic gene cluster in vivo. The success of heterologous expression of ave will facilitate Avermectins related genetic manipulations in the future. Domain swapping of combinatorial biosynthesis could be directly carried out on the recombinant BAC clones which harboring the ave cluster. An Avermectin derivatives library contains hundreds of compounds could be created based on 14F9S since every module of the Avermectins PKS could be engineered. Furthermore, it is possible to hetero-express ave in a fast growing host cell, such as E. coli, if we can modify the gene promoters and optimize the codon usage appropriately. The functional element ave could also be planted into an artificial cell factory with elaborate design directed by synthetic biology study. It will shorten the fermentation process and reduce costs in Avermectins producing.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Acknowledgments
This work was financially supported by the Ministry of Science and Technology of China (“973” Program 2013CB734003) and the National Science Foundation of China (31670030).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Barka E.A., Vatsa P., Sanchez L., Gaveau-Vaillant N., Jacquard C., Klenk H.P. Taxonomy, physiology, and natural products of actinobacteria. Microbiol Mol Biol Rev. 2015;80:1–43. doi: 10.1128/MMBR.00019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gonzalez-Ruiz A., Seaton R.A., Hamed K. Daptomycin: an evidence-based review of its role in the treatment of Gram-positive infections. Infect Drug Resist. 2016;9:47–58. doi: 10.2147/IDR.S99046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stone N.R., Bicanic T., Salim R., Hope W. Liposomal Amphotericin B (AmBisome(®)): a review of the pharmacokinetics, pharmacodynamics, clinical experience and future directions. Drugs. 2016;76:485–500. doi: 10.1007/s40265-016-0538-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies H.G., Green R.H. Avermectins and milbemycins. Nat Prod Rep. 1986;3:87–121. doi: 10.1039/np9860300087. [DOI] [PubMed] [Google Scholar]
- 5.Metcalf W.W., van der Donk W.A. Biosynthesis of phosphonic and phosphinic acid natural products. Annu Rev Biochem. 2009;78:65–94. doi: 10.1146/annurev.biochem.78.091707.100215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu L.B., Liu L.L., Yao L., Wang L.N. Efficacy and safety of tacrolimus versus cyclophosphamide for primary membranous nephropathy: a meta-analysis. Drugs. 2017;77:187–199. doi: 10.1007/s40265-016-0683-z. [DOI] [PubMed] [Google Scholar]
- 7.Cagel M., Grotz E., Bernabeu E., Moretton M.A., Chiappetta D.A. Doxorubicin: nanotechnological overviews from bench to bedside. Drug Discov Today. 2016;22:270–281. doi: 10.1016/j.drudis.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 8.Miao V., Coëffet-Legal M.F., Brian P., Brost R., Penn J., Whiting A. Daptomycin biosynthesis in Streptomyces roseosporus: cloning and analysis of the gene cluster and revision of peptide stereochemistry. Microbiology. 2005;151:1507–1523. doi: 10.1099/mic.0.27757-0. [DOI] [PubMed] [Google Scholar]
- 9.Caffrey P., Lynch S., Flood E., Finnan S., Oliynyk M. Amphotericin biosynthesis in Streptomyces nodosus: deductions from analysis of polyketide synthase and late genes. Chem Biol. 2001;8:713–723. doi: 10.1016/s1074-5521(01)00046-1. [DOI] [PubMed] [Google Scholar]
- 10.Ikeda H., Nonomiya T., Usami M., Ohta T., Omura S. Organization of the biosynthetic gene cluster for the polyketide anthelmintic macrolideavermectin in Streptomyces avermitilis. Proc Natl Acad Sci U. S. A. 1999;96:9509–9514. doi: 10.1073/pnas.96.17.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cropp T.A., Wilson D.J., Reynolds K.A. Identification of a cyclohexylcarbonyl CoA biosynthetic gene cluster and application in the production of doramectin. Nat Biotechnol. 2000;18:980–983. doi: 10.1038/79479. [DOI] [PubMed] [Google Scholar]
- 12.Wang J.B., Pan H.X., Tang G.L. Production of doramectin by rational engineering of the avermectin biosynthetic pathway. Bioorg Med Chem Lett. 2011;21:3320–3323. doi: 10.1016/j.bmcl.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 13.Gaisser S., Kellenberger L., Kaja A.L., Weston A.J., Lill R.E., Wirtz G. Direct production of ivermectin-like drugs after domain exchange in the avermectin polyketide synthase of Streptomyces avermitilis ATCC31272. Org Biomol Chem. 2003;1:2840–2847. doi: 10.1039/b304022d. [DOI] [PubMed] [Google Scholar]
- 14.Li M., Chen Z., Lin X., Zhang X., Song Y., Wen Y. Engineering of avermectin biosynthetic genes to improve production of ivermectin in Streptomyces avermitilis. Bioorg Med Chem Lett. 2008;18:5359–5363. doi: 10.1016/j.bmcl.2008.09.061. [DOI] [PubMed] [Google Scholar]
- 15.Kim M.S., Cho W.J., Song M.C., Park S.W., Kim K., Kim E. Engineered biosynthesis of milbemycins in the avermectin high-producing strain Streptomyces avermitilis. Microb Cell Fact. 2017;16(1):9. doi: 10.1186/s12934-017-0626-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bierman M., Logan R., O'Brien K., Seno E.T., Rao R.N., Schoner B.E. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 17.Flett F., Mersinias V., Smith C.P. High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol Lett. 1997;155:223–229. doi: 10.1111/j.1574-6968.1997.tb13882.x. [DOI] [PubMed] [Google Scholar]
- 18.Bibb M.J., Buttner M.J., Chater K.F., Hopwood D.A. John Innes Foundation; 2000. Practical Streptomyces genetics. [Google Scholar]
- 19.Shizuya H., Birren B., Kim U.J., Mancino V., Slepak T., Tachiiri Y. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci U. S. A. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi X., Zeng H., Xue Y., Luo M. A pair of new BAC and BIBAC vectors that facilitate BAC/BIBAC library construction and intact large genomic DNA insert exchange. Plant Methods. 2011;7:33. doi: 10.1186/1746-4811-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang L., Chen S., Vergin K.L., Giovannoni S.J., Chan S.W., DeMott M.S. DNA phosphorothioation is widespread and quantized in bacterial genomes. Proc Natl Acad Sci U. S. A. 2011;108:2963–2968. doi: 10.1073/pnas.1017261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L., Chen S., Xu T., Taghizadeh K., Wishnok J.S., Zhou X. Phosphorothioation of DNA in bacteria by and genes. Nat Chem Biol. 2007;3:709–710. doi: 10.1038/nchembio.2007.39. [DOI] [PubMed] [Google Scholar]
- 23.Zhou X., He X., Liang J., Li A., Xu T., Kieser T. A novel DNA modification by sulphur. Mol Microbiol. 2005;57:1428–1438. doi: 10.1111/j.1365-2958.2005.04764.x. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J.F., Russell D.W. third ed. Cold Spring Harbor Laboratory Press; 2001. Molecular cloning: a laboratory manual. [Google Scholar]
- 25.Luo M., Wang Y.H., Frisch D., Joobeur T., Wing R.A., Dean R.A. Melon bacterial artificial chromosome (BAC) library construction using improved methods and identification of clones linked to the locus conferring resistance to melon Fusarium wilt (Fom-2) Genome. 2001;44:154–162. [PubMed] [Google Scholar]
- 26.Burg R.W., Miller B.M., Baker E.E., Birnbaum J., Currie S.A., Hartman R. Avermectins, new Family of potent anthelmintic agents: producing organism and fermentation. Antimicrob Agents Chemother. 1979;15:361–367. doi: 10.1128/aac.15.3.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen L., Li P., Deng Z., Zhao C. Ornithine Transcarbamylase ArgK plays a dual role for the self-defense of phaseolotoxin producing Pseudomonas syringae pv. phaseolicola. Sci Rep. 2015;5:12892. doi: 10.1038/srep12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ikeda H., Omura S. Control of avermectin biosynthesis in Streptomyces avermitilis for the selective production of a useful component. J Antibiot (Tokyo) 1995;48:549–562. doi: 10.7164/antibiotics.48.549. [DOI] [PubMed] [Google Scholar]
- 29.Zhuo Y., Zhang T., Wang Q., Cruz-Morales P., Zhang B., Liu M. Synthetic biology of avermectin for production improvement and structure diversification. Biotechnol J. 2014;9:316–325. doi: 10.1002/biot.201200383. [DOI] [PubMed] [Google Scholar]
- 30.Zhuo Y., Zhang W., Chen D., Gao H., Tao J., Liu M. Reverse biological engineering of hrdB to enhance the production of avermectins in an industrial strain of Streptomyces avermitilis. Proc Natl Acad Sci U. S. A. 2010;107(25):11250–11254. doi: 10.1073/pnas.1006085107. [DOI] [PMC free article] [PubMed] [Google Scholar]