Fig. 6.
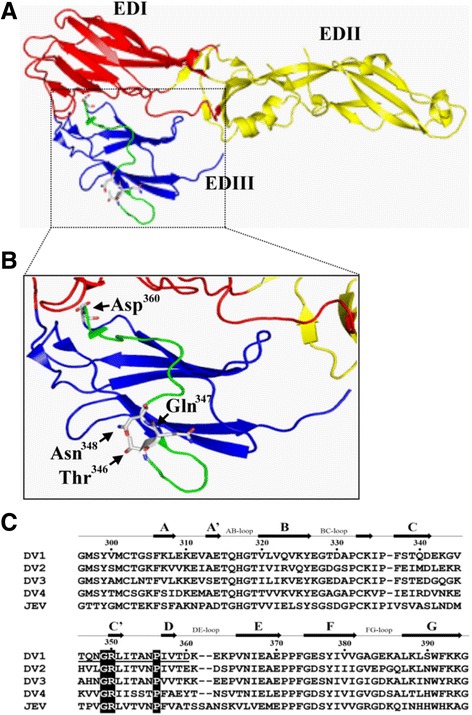
Overview of the DV envelope protein and epitope localization of anti-E mAbs. a Crystal structure of the post-fusion DV1-soluble fragment E (sE) (PDB accession number 3G7T), with subunits highlighted and colored by domains (EDI, red; EDII, yellow; EDIII, blue). The location of the mAb epitope (residues 346–360) is highlighted by green. b Enlargement of the EDIII region (blue) with mAb epitope region (green). The major residues of the loop of mAb epitope are marked, the starting residue Thr346, and three contact residues Gln347, Asn348, and Asp360. c Amino acid sequence alignment of four dengue serotypes EDIII and JEV EDIII. The sequences of EDIII from DV1 (strain Hawaii), DV2 (NGC strain), DV3 (H87), DV4 (H241), and JEV (T1P1) are shown. Conserved amino acid residues within the epitope region are highlighted by black blocks. Secondary structure elements of DV1 EDIII are shown with arrows (β-strands) and are labeled according to Shresth et al. [29]. Residue numbers correspond to the full-length DV1 E protein. Amino acid residues marked by underlining indicate the sequence of the epitope
