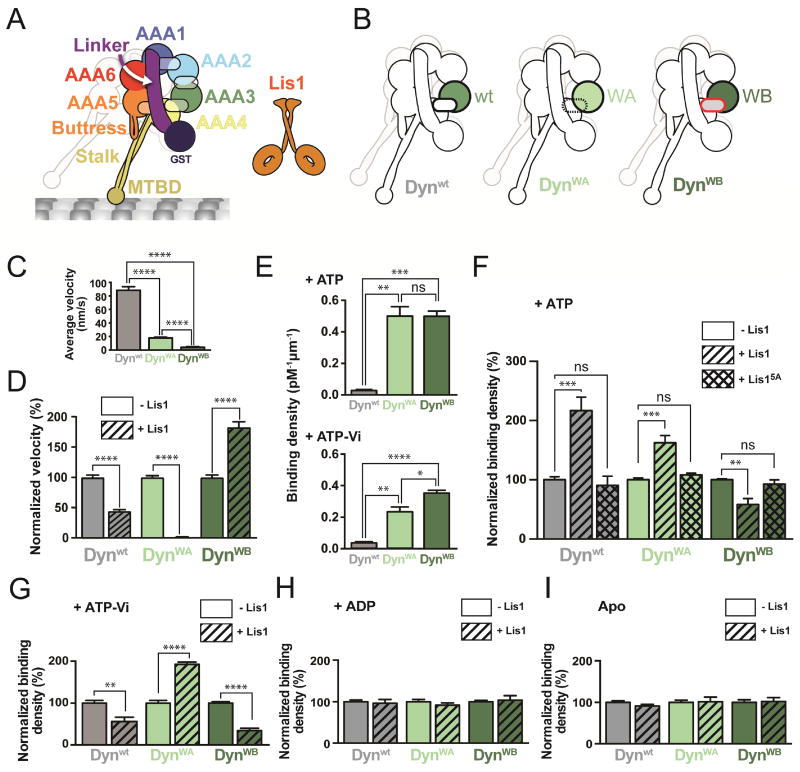
Figure 1. Lis1 has two modes of regulating dynein.
(A) Schematic of the dynein construct used in this study. The semi-transparent ovals represent the nucleotide-binding sites in AAA1-4. The second protomer is faded out for clarity. The β-propellers in the Lis1 dimer (orange) are represented as rings. (B) AAA3 variants used in this study: Dynwt (wild type AAA3), DynWA (Walker A mutation in AAA3), and DynWB (Walker B mutation in AAA3). (C) Average velocities of dynein variants (n>103 events per data point). See Figure S1B for representative kymographs. (D) Normalized average velocities of dynein variants in the absence (solid bars) and presence (hatched bars) of 300 nM Lis1 (n>103 events per data point). Velocities were normalized by setting those in the absence of Lis1 [from (C)] to 100%. (E) Binding densities of dynein variants in the presence of ATP (top) or ATP-Vi (bottom) (n>8 fields of view per data point). See also Table S1. (F) Normalized binding densities of dynein variants alone (solid bars), or in the presence of 300 nM Lis1 (hatched bars), or 300 nM Lis15A (cross-hatched bars) in the presence of ATP. Binding densities were normalized by setting those in the absence of Lis1 [from (E), top] to 100% (n=12 fields of view per data point). (G–I) Normalized binding densities of dynein variants alone (solid bars), or in the presence of 300 nM Lis1 (hatched bars) in the presence of 1 mM ATP-Vi (G), 1 mM ADP (H) and 2.5 units/mL apyrase (“Apo”) (I) (n=8 fields of view per data point). Normalized binding densities in the absence of Lis1 shown in (G) are those in (E, bottom) without normalization. Statistical significance was calculated using an unpaired t-test with Welch’s correction for both velocity (C and D) and binding density (E–I). P-values: ns, not significant; *, <0.05; **, <0.01; ***, <0.001, ****, <0.0001. Data are shown as mean and standard error of mean.