Significance
α-Synuclein (aSyn) plays a key role in neurodegenerative disorders known as “synucleinopathies.“ By tracking individual primary neurons overexpressing wild-type and mutant aSyn versions we have determined how pathological hallmarks like alterations in aSyn expression and stability, aggregation, or serine 129 phosphorylation (PS129) contribute to neuronal death. We have found that the E46K mutant displayed the highest toxicity. Neurotoxicity was independent of aggregation but correlated with PLK2-dependent PS129. Surprisingly, inhibiting PS129 did not affect neuronal survival, suggesting that this modification is an epiphenomenon. Finally, we set up an assay to score for cell-autonomous and nonautonomous toxicity. While we identified a minor non–cell-autonomous component restricted to proximal neurons, E46K toxicity was mostly cell autonomous.
Keywords: alpha-synuclein, E46K mutation, serine 129 phosphorylation, neuronal death, autonomous toxicity
Abstract
α-Synuclein (aSyn) is the main driver of neurodegenerative diseases known as “synucleinopathies,” but the mechanisms underlying this toxicity remain poorly understood. To investigate aSyn toxic mechanisms, we have developed a primary neuronal model in which a longitudinal survival analysis can be performed by following the overexpression of fluorescently tagged WT or pathologically mutant aSyn constructs. Most aSyn mutations linked to neurodegenerative disease hindered neuronal survival in this model; of these mutations, the E46K mutation proved to be the most toxic. While E46K induced robust PLK2-dependent aSyn phosphorylation at serine 129, inhibiting this phosphorylation did not alleviate aSyn toxicity, strongly suggesting that this pathological hallmark of synucleinopathies is an epiphenomenon. Optical pulse-chase experiments with Dendra2-tagged aSyn versions indicated that the E46K mutation does not alter aSyn protein turnover. Moreover, since the mutation did not promote overt aSyn aggregation, we conclude that E46K toxicity was driven by soluble species. Finally, we developed an assay to assess whether neurons expressing E46K aSyn affect the survival of neighboring control neurons. Although we identified a minor non–cell-autonomous component spatially restricted to proximal neurons, most E46K aSyn toxicity was cell autonomous. Thus, we have been able to recapitulate the toxicity of soluble aSyn species at a stage preceding aggregation, detecting non–cell-autonomous toxicity and evaluating how some of the main aSyn hallmarks are related to neuronal survival.
The abnormal accumulation of the presynaptic protein α-synuclein (aSyn) is the hallmark of a broad spectrum of neurodegenerative disorders collectively known as “synucleinopathies” (1). Mutations in the gene encoding aSyn (SNCA) are associated with familial forms of these diseases. For example, nonsynonymous point mutations and triplications of the SNCA gene are linked to autosomal dominant Parkinson’s disease (PD), while gene duplication of the SNCA gene is sufficient to cause dementia with Lewy bodies (DLB) (2). Indeed, genome-wide association studies have identified the SNCA gene as one of the strongest risk loci in sporadic forms of PD (3, 4). Polymorphisms within the SNCA promoter increase PD susceptibility (5), and a PD-associated risk variant located in a distal enhancer of the SNCA gene regulates aSyn expression (6). Thus, aSyn would appear to play a central role in these pathologies, and enhanced aSyn expression may even be sufficient to cause familial forms of synucleinopathies and favor the onset of sporadic cases.
The molecular mechanisms by which aSyn causes neuronal death remain elusive. The aSyn protein is intrinsically disordered, existing predominantly as a monomer (7), although tetrameric conformations could exist in a dynamic equilibrium (8). A misfolded conformation of aSyn monomers could be more prone to assemble into intermediate or oligomeric aSyn species, triggering fibrillization and their final aggregation into Lewy bodies (LBs), the pathological hallmark of synucleinopathies (9–11). The enhanced expression of WT aSyn may be sufficient to initiate this aggregation cascade. Similarly, pathological point mutations could contribute to aSyn aggregation by impairing protein degradation, thereby augmenting the steady-state protein levels and/or favoring the accumulation of aggregation-prone aSyn conformations. In vitro and in vivo studies suggest that aSyn mutations alter degradation pathways (i.e., chaperone-mediated autophagy) (12) and that they modulate aSyn aggregation (13). However, there is still no conclusive evidence about which are the toxic aSyn species (monomers, oligomers, or fibrillar/aggregated LBs) or how these toxic species contribute to neurodegeneration. When forming LBs, most aSyn is phosphorylated at serine 129 (S129) (14), a modification that might promote aggregation or that might occur after aSyn assembles into LBs. Despite much effort in cellular and animal models, it remains unclear how this hallmark relates to neurodegeneration (15).
In recent years, the hypothesis that pathological proteins spread from neuron to neuron has been proposed as a non–cell-autonomous mechanism to explain the progression of PD neurodegeneration. Toxic aSyn protein species could be secreted by cells and taken up by surrounding neurons, serving as a template to trigger the misfolding and aggregation of the endogenous protein in a prion-like manner (16). Consistent with this idea, aSyn of CNS origin has been detected in the cerebrospinal fluid of healthy individuals and persons with PD (17, 18). Overexpression of aSyn in neuronal models promotes aSyn release into the extracellular milieu (19, 20), and exogenous preformed aSyn fibrils injected into mouse brains can be taken up by neurons, promoting the aggregation of endogenous aSyn in these cells (21–24). Moreover, brain propagation with nonfibrillar aSyn protein species has also been observed in mice (25).
The spreading of toxic aSyn species could explain the ascending pattern of LB distribution in PD postmortem human brains: from the lower brainstem toward the pons, mesencephalon, and to the cortical regions at later stages of the disease (26). While this hypothesis has generated much interest, the involvement of aSyn-dependent, cell-autonomous mechanisms targeting neuronal populations at different stages during disease progression as a result of differential vulnerability has not been ruled out (27, 28). Understanding the extent to which an aSyn pathology is caused by cell-autonomous as opposed to non–cell-autonomous mechanisms is a fundamental issue when attempting to untangle the events that underpin the progression of these pathologies.
While different animal and cellular models recapitulate different aspects of aSyn pathology, such as aggregation or phosphorylation, they do not allow the relevance of such features to be linked to neuronal survival. To address this limitation, we previously developed an automated microscopy method to track individual neurons expressing fluorescently tagged neurotoxic proteins over long periods of time. Longitudinal tracking of individual neurons enables the risk of neuronal death to be determined quantitatively and Cox regression models to be applied to evaluate predictive factors of neuronal death (29–33). Given the relevance of neurotoxic protein dynamics in neurodegeneration, this approach was then adapted to determine the protein’s half-life in individual living neurons (optical pulse labeling, OPL) (34, 35). Based on this approach, we have now developed a neuronal model to assess the risk of death when neurons express WT or mutant aSyn alleles. We then investigated whether the main hallmarks of aSyn pathology, such as protein stability, aggregation, or S129 phosphorylation, contribute to neuronal death. Finally, we developed an assay to quantitatively assess whether aSyn toxicity occurs in a cell-autonomous or a non–cell-autonomous manner, based on the capacity of aSyn-expressing neurons to affect the survival of surrounding cells.
Results
The E46K aSyn Pathological Mutation Is the Most Toxic Mutation in Primary Cultures of Rat Cortical Neurons.
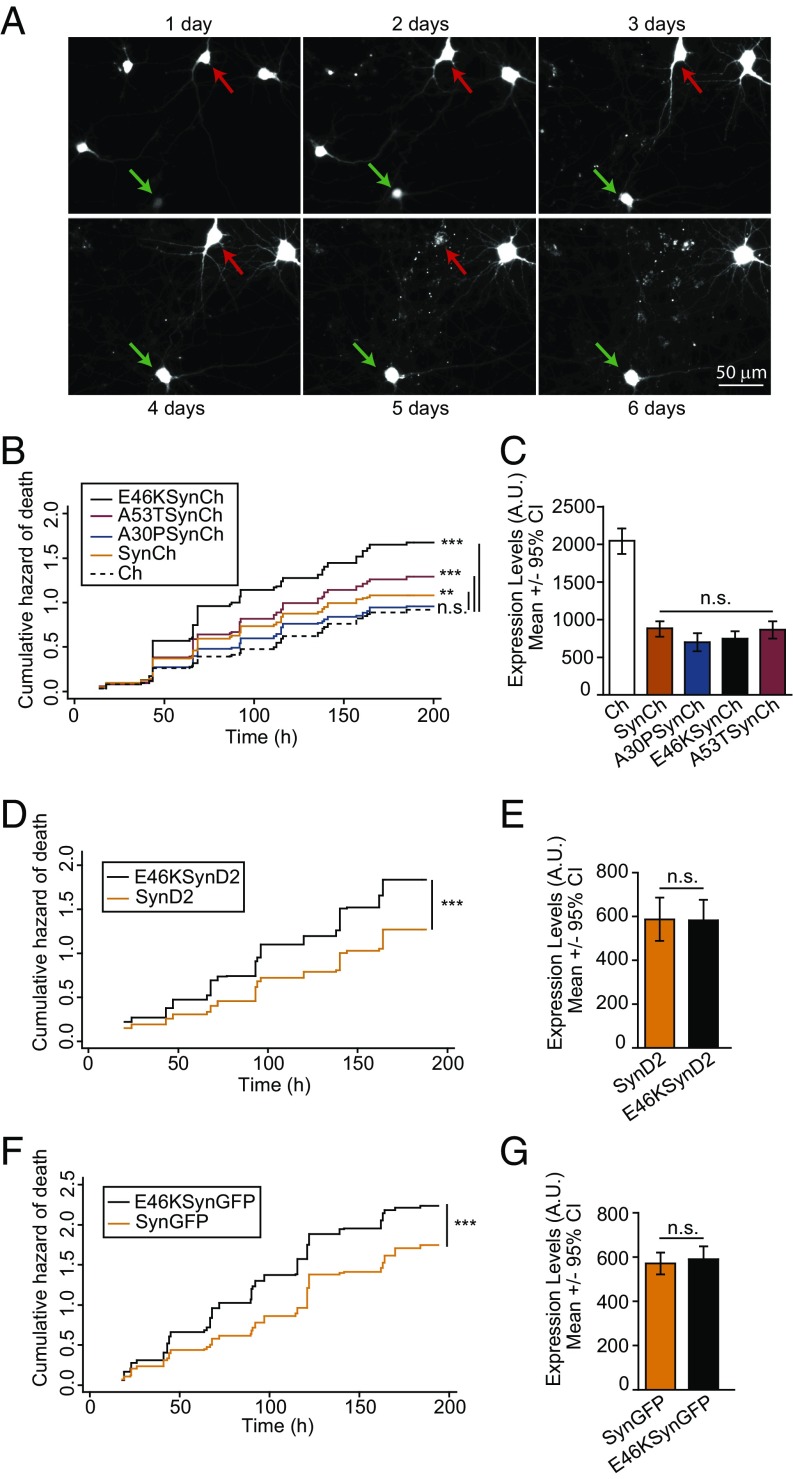
To identify and track aSyn-expressing neurons over time, we generated WT and pathological aSyn alleles (A30P, E46K, and A53T) fused to monomeric Cherry (Ch) fluorescent protein. The fusions were inserted at the C terminus to avoid interference with any aSyn N-terminal posttranslational modifications (36). Individual rat cortical primary neurons transfected with plasmids encoding Ch-tagged WT or mutant aSyn versions (SynCh, A30PSynCh, E46KSynCh, and A53TSynCh) and the Ch protein alone as a control were subjected to longitudinal tracking by automated microscopy and survival analysis (Fig. 1A). Both fluorescence intensity (as a surrogate measure for protein expression) (Fig. S1) and survival time (the last time each neuron was observed alive) were estimated for individual neurons. A Cox proportional hazard (CPH) analysis indicated that the WT and mutant (A53T and E46K) aSyn significantly increased the risk of death with respect to control (Ch) neurons. The E46K mutant was the most toxic construct (Fig. 1B) based on the hazard ratio (HR) coefficients for each of the aSyn constructs relative to the control (Ch): E46KSynCh, HR 1.82, P < 0.001, 95% CI 1.6–2.1; A53TSynCh, HR 1.36, P < 0.001, 95% CI 1.2–1.6; SynCh, HR 1.22, P < 0.01, 95% CI 1.1–1.4; A30PSynCh, HR 1.1, P = 0.46, 95% CI 0.9–1.3. The similar steady-state protein levels of all aSyn variants tested ruled out any differences in protein abundance underlying the distinct toxicity observed among the aSyn alleles (Fig. 1C). Moreover, similar results were obtained when using monomeric GFP- or Dendra2 (D2)-tagged constructs (Fig. 1 D–G): E46KSynCh versus SynCh, HR 1.46, P < 0.001, 95% CI 1.35–1.58; E46KSynD2 versus SynD2, HR 1.39, P < 0.001, 95% CI 1.2–1.6; E46KSynGFP versus SynGFP, HR 1.35, P < 0.001, 95% CI 1.2–1.5. Notably, the different fluorescent tags used in this study produced different levels of background toxicity (Table S1 and Fig. S2A), and therefore the intrinsic D2 and GFP toxicity masked the WT aSyn toxicity (Fig. S2 C and E). To rule out the possibility that tagging could affect aSyn behavior/toxicity, we tested the toxicity of untagged aSyn versions expressed from a dicistronic mRNA, which expresses GFP in an internal ribosome entry site (IRES)-dependent manner. Since IRES-driven GFP fluorescence was not sufficient to identify and track transfected neurons, untagged constructs (pCAGGs-Syn-IRES-GFP or pCAGGs-E46KSyn-IRES-GFP) were cotransfected with Ch (Fig. S3). Similar toxicity levels were found between untagged and tagged aSyn versions (Fig. S3 A and B). Taken together, our analyses identify E46K aSyn as the most toxic pathological mutation.
Fig. 1.
The E46K aSyn mutant significantly increases the risk of neuronal death in primary cultures of rat cortical neurons. (A) Longitudinal tracking with automated microscopy of individual primary rat cortical neurons transiently transfected with Ch. The green arrow points to a neuron tracked longitudinally for up to 6 dpt; the red arrow indicates a neuron that died 5 dpt. (B) Cumulative hazard estimates of primary neurons transfected with WT aSyn (SynCh), with the pathological mutants A30PSynCh, E46KSynCh, or A53TSynCh, or with Ch as control. Results show CPH analysis of 500–750 neurons per condition from four independent experiments. (C) Ch fluorescence intensity of individual neurons 20–24 h after transfection (n = 80–160 neurons per condition from a representative experiment; Kruskal–Wallis and Dunn’s post hoc test). (D and F) Cumulative hazard estimates of primary neurons transfected with D2/GFP-tagged WT aSyn or the E46K mutant (SynD2, SynGFP, E46KSynD2, E46KSynGFP). CPH analysis: D2 fusions, n = around 500 neurons per condition from four independent experiments; GFP fusions, n = around 900 neurons per condition from five independent experiments. (E and G) Expression of aSyn (WT or E46K mutant) in neurons at 20–24 h after transfection (n = around 100 neurons per condition from representative experiments Mann–Whitney test). All error bars indicate 95% CIs; n.s., nonsignificant; **P < 0.01; ***P < 0.001.
Fig. S1.
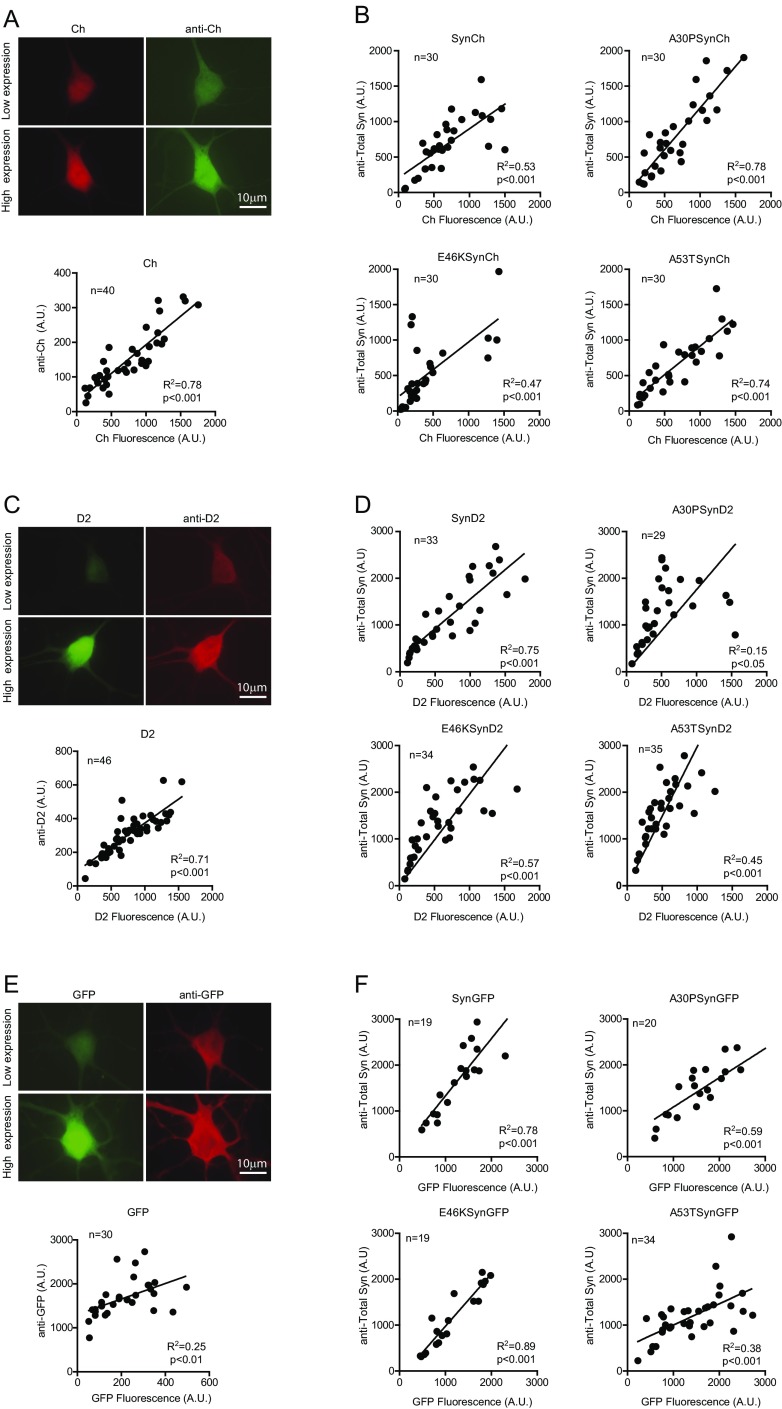
Fluorescence intensity of aSyn Ch/D2/GFP-tagged constructs as a surrogate for aSyn protein levels. (A, C, and E) Rat cortical neurons transfected with the Ch (A), D2 (C), and GFP plasmids (E) and immunostained with anti-Ch–, anti-D2–, and anti-GFP–specific antibodies. The fluorescence intensity of Ch, D2, and GFP and of the secondary antibodies recognizing anti-Ch, anti-D2, and anti-GFP was quantified in individual neurons. (B, D, and F) Neurons were transfected with Ch/D2/GFP-tagged versions of aSyn, and immunostaining was performed with an anti-aSyn–specific antibody (anti-Total aSyn). Fluorescence intensity from Ch, D2, and GFP and from secondary antibodies recognizing the anti-Total aSyn was quantified in individual neurons. Coefficient of determination (R2) and P values were estimated by correlation analysis.
Table S1.
CPH model of the effect of fluorescent reporter expression levels at 18 h posttransfection on neuronal survival
| Group | HR | 95% CI | P value |
| D2 (n = 212) | 1.0003 | 1.0002–1.0004 | <0.001*** |
| GFP (n = 220) | 1.0016 | 1.0009–1.0024 | <0.001*** |
| Ch (n = 225) | 0.9998 | 0.9996–1.0000 | 0.085 |
n, number of neurons.
Fig. S2.
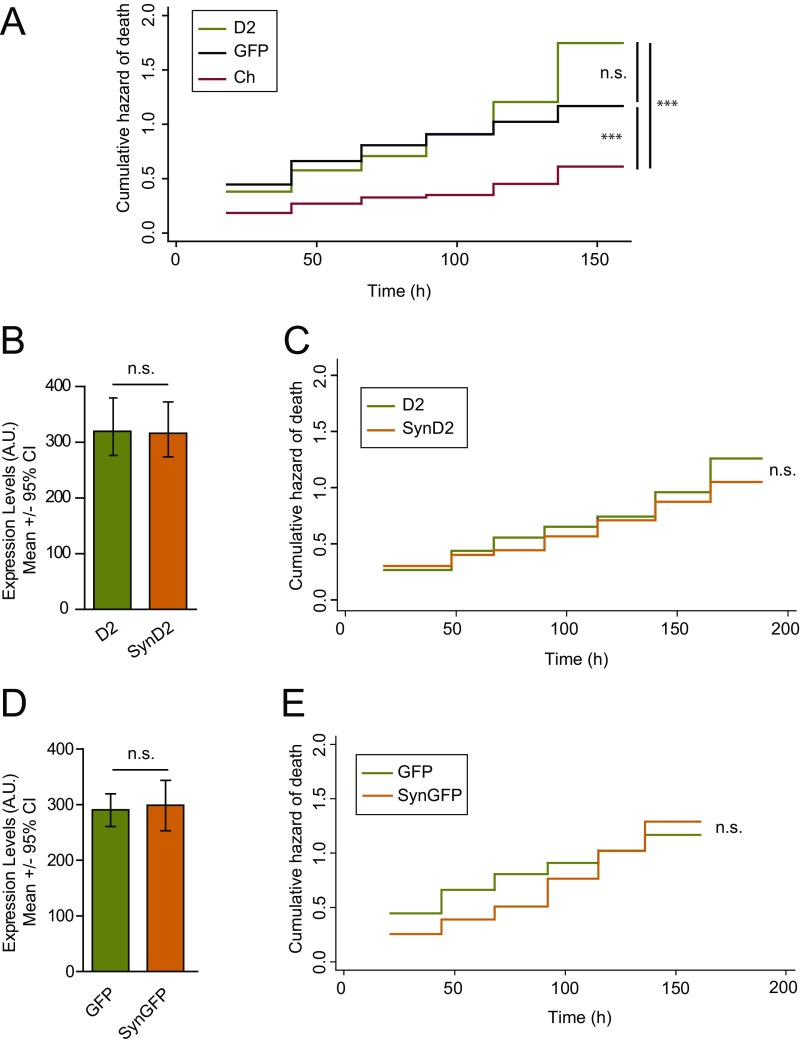
Ch, D2, and GFP fluorescent proteins affect the survival of cortical neurons distinctly. (A) Cumulative risk estimates of primary neurons transfected with Ch, D2, and GFP (D2 = 386, GFP = 210, and Ch = 141 neurons, respectively; log-rank test; n.s., nonsignificant, ***P < 0.001). (B) Cohorts of neurons expressing similar levels of D2 and SynD2 were selected for further analysis of the risk of neuronal death (D2 = 119 and SynD2 = 125 neurons, respectively; Mann–Whitney test; n.s., nonsignificant). (C) The risk of death of cohorts of neurons expressing similar levels of WT aSyn (SynD2) and control protein D2 (from B) was compared (log-rank test; n.s., nonsignificant). (D) Cohorts of neurons expressing similar levels of GFP and SynGFP were selected (SynGFP = 132 and GFP = 210 neurons; Mann–Whitney test; n.s., nonsignificant). (E) The risk of death in cohorts of neurons was compared after expressing similar levels of WT aSyn (SynGFP) and control protein GFP (from D; log-rank test; n.s., nonsignificant). All error bars represent the 95% CIs.
Fig. S3.
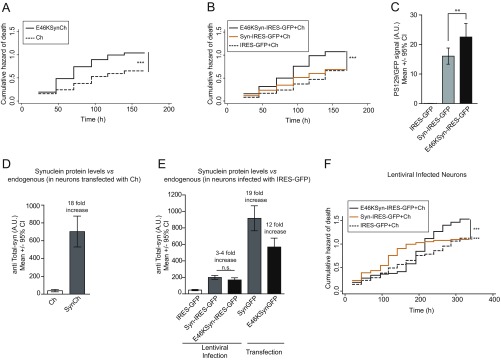
Untagged IRES-GFP versions of aSyn show neuronal toxicity similar to that of Ch-tagged versions. (A) Cumulative hazard estimates of primary rat cortical neurons coexpressing E46KSynCh and Ch as control (n = 250–300 neurons per condition; CPH analysis; ***P < 0.001). (B) Cumulative hazard of death of primary rat cortical neurons coexpressing WT or E46K aSyn (Syn-IRES-GFP, E46KSyn-IRES-GFP) and Ch. Control conditions are neurons coexpressing IRES-GFP and Ch (n = 250–300 neurons per condition; CPH analysis; ***P < 0.001). (C) Quantification of anti-PS129 binding relative to GFP (from IRES-dependent expression) in individual primary rat cortical neurons expressing IRES-GFP, Syn-IRES-GFP, and E46KSyn-IRES-GFP from immunofluorescence experiments (n = 42–66 neurons per condition; one-way ANOVA and Bonferroni’s post hoc test; **P < 0.01). All error bars represent 95% CIs. (D) Quantification of total aSyn protein levels in immunostained individual neurons transfected with Ch and SynCh. Total aSyn levels in Ch-transfected neurons are indicative of endogenous aSyn levels (n = 10–12 neurons per condition). (E) Quantification of anti-total aSyn binding in individual neurons infected with lentivirus (IRES-GFP as control, Syn-IRES-GFP, or E46KSyn-IRES-GFP) or transfected with SynGFP and E46KSynGFP. Total aSyn levels in IRES-GFP–infected neurons are indicative of endogenous aSyn levels (n = 20–70 neurons per condition; Kruskal–Wallis and Dunn’s post hoc test). All error bars represent 95% CIs. (F) Longitudinal survival analysis in primary rat cortical neurons infected with aSyn-expressing lentivirus. Cumulative hazard estimates of primary rat cortical neurons transfected with Ch and infected 4 h later with E46KSyn-IRES-GFP, Syn-IRES-GFP, or IRES-GFP, respectively (n = 150–300 neurons per condition; CPH analysis; ***P < 0.001).
The E46K Mutation Does Not Affect Either aSyn Protein Turnover (Half-Life) or Its Propensity to Aggregate.
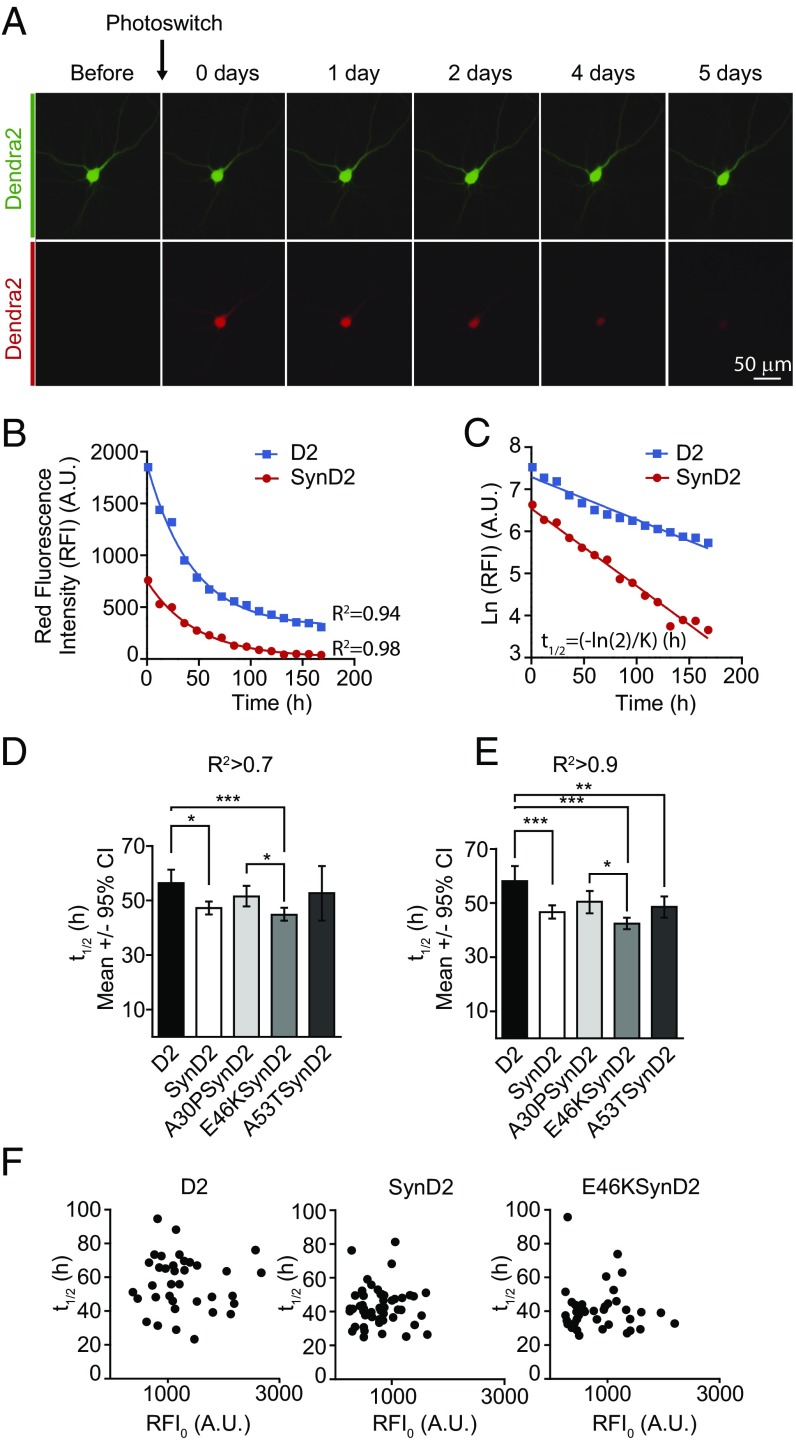
A possible explanation for the contribution of pathological aSyn mutations to neurodegeneration is that aSyn mutations may interfere with protein-degradation pathways (12, 37), thereby increasing aSyn concentrations and toxicity. Alternatively, the aSyn mutations may favor the adoption of more toxic, aggregation-prone protein conformations (13). Thus, we investigated whether E46K-dependent toxicity was associated with altered protein stability and/or degradation. We performed OPL experiments to estimate protein turnover (half-life) in primary neurons in vivo (34, 35). The OPL approach overcomes the need for radioactivity and the use of translational inhibitors in conventional protocols, and it enables protein stability to be studied in individual neurons. OPL experiments are based on the biophysical properties of the fluorescent protein D2 (38), a photoconvertible fluorescent protein that normally emits green fluorescence but that changes its conformation irreversibly upon an intense pulse with blue light, emitting red fluorescence. The photoconverted protein pool can be tracked over time in vivo using automated longitudinal microscopy, such that the red fluorescence intensity (RFI) will decline over time due to protein turnover (Fig. 2A). RFI decays following an exponential pattern in individual neurons (Fig. 2B). Thus, logarithmic transformation of RFI measurements for each individual neuron and further adjustment by linear regression enables the D2 half-life in individual neurons to be determined from the slope (K): t1/2 = −ln (2)/K (Fig. 2C).
Fig. 2.
The E46K mutation does not alter aSyn protein stability. (A) Protein turnover (half-life) estimated in living neurons by OPL. (Upper Row) Illumination of a D2-transfected neuron with light at a wavelength of 488 nm induces the emission of green fluorescence. (Lower Row) After a brief pulse with intense blue light (photoswitching), a proportion of the D2 molecules photoconvert and emit red fluorescence. Green and red fluorescence intensities in single neurons are longitudinally monitored by automated microscopy. (B) Example of two photoconverted neurons in which changes in RFI over time fit an exponential decay. (C) Logarithmic transformation of the intensity of RFI [ln(RFI)] and adjustment by linear regression. The slope (K) enables the D2 half-life to be estimated (t1/2 = −ln (2)/K) in individual neurons in hours (h). (D and E) Half-life estimated for D2-tagged WT aSyn (SynD2) and pathological mutants A30PSynD2, E46KSynD2, and A53TSynD2 in neurons subjected to OPL. Graphs show the estimated half-life for neurons with coefficients of determination R2 >0.7 (D) and R2 >0.9 (E) (Kruskal–Wallis and Dunn’s post hoc test, n = 130–290 neurons per condition from five independent experiments). (F) No correlation was found between the initial RFI after photoconversion (RFI0) and the protein half-life estimated in individual neurons transfected with D2 or D2 aSyn-tagged constructs (n = 30–50 neurons per condition, a representative experiment). All error bars indicate 95% CIs; *P < 0.05, **P < 0.01, ***P < 0.001.
The stability of D2-tagged WT and mutant aSyn alleles expressed in primary cortical neurons was assessed by OPL. The half-life of WT aSyn was previously estimated to be around 50 h in nonneuronal and neuronal cell lines (7, 39). As such, images were obtained every 12 h after photoconversion over a total of ≈160 h. In the majority of neurons (around 95%) the decay of WT or mutant aSyn-D2 protein followed exponential decay kinetics, such that the half-life was estimated only in neurons with a coefficient of R2 >0.7. Accordingly, we found the half-life of WT aSyn in primary neurons to be ≈47 h. In contrast to earlier reports (12), we did not observe a longer half-life for the A53T or A30P aSyn mutants compared with the WT (Fig. 2D). Moreover, and despite its higher toxicity, the E46K mutation did not affect aSyn protein turnover. Similar results were obtained when only neurons with a coefficient of determination R2 >0.9 were analyzed (≈78% of the total neurons) (Fig. 2E). Notably, the estimated half-life in these experiments did not depend on the initial amount of photoswitched protein (initial RFI, RFI0) (Fig. 2F). Thus, the toxicity observed with E46K mutation was not due to an increase in protein stability.
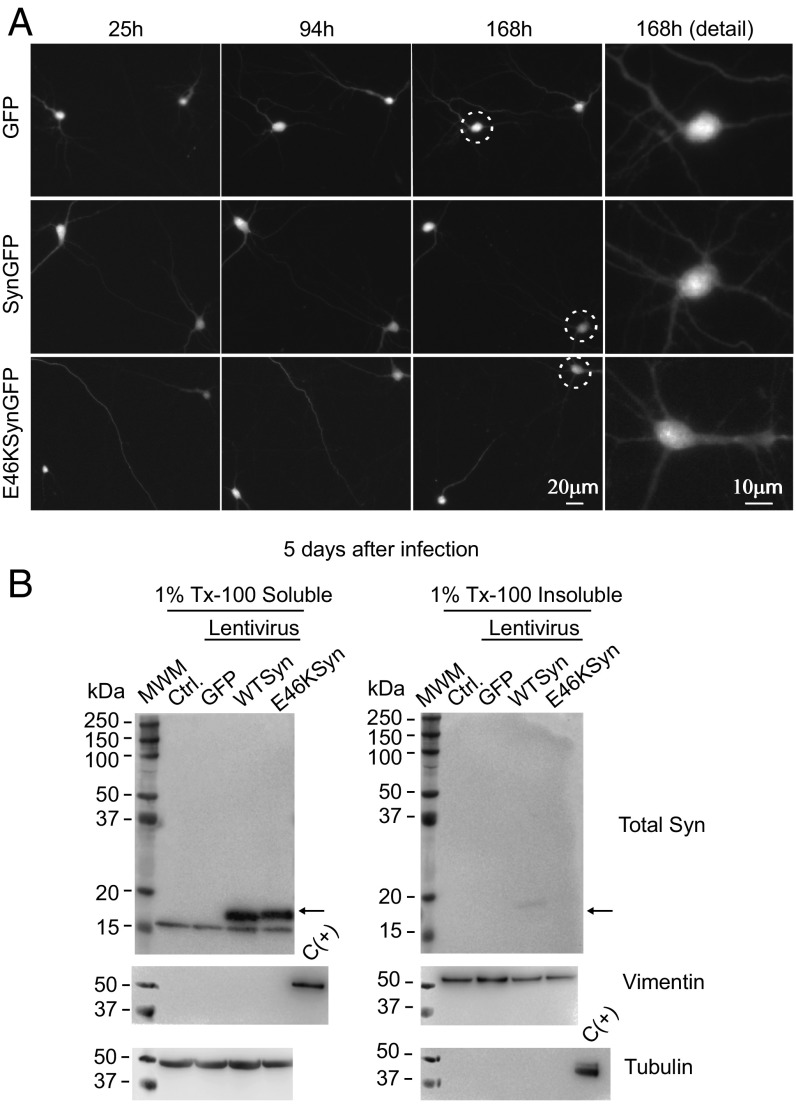
We investigated whether E46K toxicity resulted from enhanced abnormal aggregation, based on the formation of round, bright puncta in neurons expressing fluorescently tagged toxic proteins (e.g., mutant huntingtin) constituting visual proof of aggregation (30, 32, 33, 35). Visual inspection of cortical primary neurons transfected with WT or E46K aSyn revealed that aSyn was distributed homogeneously, and the formation of aggregates was not apparent during the experiments (see representative images in Fig. 3A). Since aSyn aggregates are resistant to extraction with nonionic detergents such as 1% Triton X-100 (1% Tx-100) (40), we performed detergent-extraction experiments to assess whether E46K aSyn had a greater propensity to form aggregates. We infected primary cortical neurons with lentiviruses expressing WT or E46K together with an IRES-driven GFP reporter (41). Compared with transient transfection experiments, lentiviral transduction yielded lower aSyn levels in individual neurons and recapitulated E46K and WT aSyn toxicity (Fig. S3 E and F). Since our longitudinal survival experiments were carried out for up to 8 d, extracts of the primary neuronal cultures were prepared 5 and 10 d after lentiviral infection by homogenization in buffer containing 1% Tx-100, and the Tx-100–soluble and –insoluble protein fractions were analyzed by Western blots. If aggregate formation occurs, aSyn should appear in the insoluble protein fraction, but both WT and E46K aSyn were found predominantly in the Tx-100–soluble fraction at 5 and 10 d after infection, with a mass of ≈16 kDa (Fig. 3B and Fig. S4). As controls for correct extract fractionation, tubulin appeared in the soluble fraction and vimentin in the insoluble fraction (42). Thus, E46K toxicity cannot be explained by changes in aSyn stability or aggregation. Indeed, our model recapitulates the induction of E46K aSyn-dependent neuronal toxicity before insoluble aggregates appear. Accordingly, we postulate that soluble aSyn species may act as toxicity drivers and that aggregation may take place at later stages of neurodegeneration.
Fig. 3.
The E46K mutant does not form Tx-100–insoluble aggregates in rat primary cortical neurons during a longitudinal survival experiment. (A) Visual inspection does not reveal aggregate formation in neurons transfected with GFP, SynGFP, or E46KSynGFP. The circles show neurons that are amplified in the image at the Right. (B) Rat cortical primary neurons were infected with lentivirus expressing aSyn (WT or E46K mutant) together with an IRES-driven GFP reporter (41), and 5 d after lentiviral infection the primary neuronal cultures were extracted with 1% Tx-100. The detergent-soluble and -insoluble protein fractions were separated by ultracentrifugation and analyzed by Western blots. As reported elsewhere (42), tubulin appears in the soluble fraction and vimentin in the insoluble fraction. A sample from the opposite fraction was used as a positive control [C(+)]. The aSyn from these cells is predominantly detected in the 1% Tx-100–soluble fraction, with a mass of ≈16 kDa. Experiments were performed twice with similar results. Ctrl, control noninfected neurons; MWM, molecular weight marker.
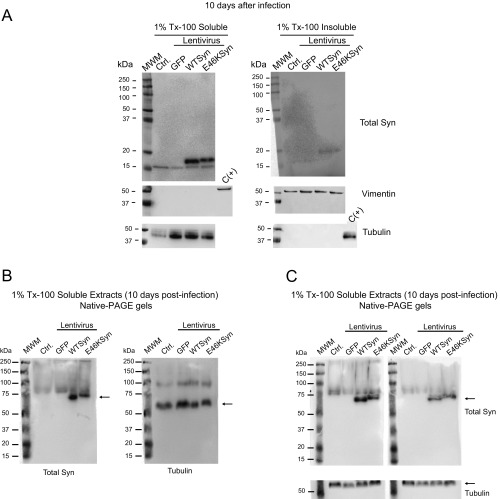
Fig. S4.
The E46K aSyn pathological mutation does not form Tx-100–insoluble aggregates in primary rat cortical neurons 10 d after lentiviral infection. (A) Ten days after lentiviral infection, extracts of primary neuron cultures were homogenized in 1% Tx-100, and the detergent-soluble and -insoluble protein fractions obtained by ultracentrifugation at 100,000 × g were analyzed in Western blots. The aSyn is predominantly detected in the Tx-100–soluble fraction with a mass of ≈16 kDa; as reported, tubulin appears in the soluble fraction, and vimentin appears in the insoluble fraction. A sample from the opposite fraction was used as a positive control [C(+)]. Each experiment was performed twice with similar results. (B) Analysis of 1% Tx-100–soluble fractions from neuronal protein extracts infected with aSyn expressing lentivirus by native PAGE gels and Western blot. Rat cortical primary neurons were infected with lentivirus expressing WT or E46K mutant aSyn. Ten days after lentiviral infection, extracts of primary neuron cultures were homogenized with 1% Tx-100. The detergent-soluble fraction was separated by ultracentrifugation, run in native PAGE gels, and analyzed by Western blot with a specific antibody against Total aSyn. aSyn is detected with an apparent mass between 60–70 kDa. The same membrane was stripped and incubated with anti-tubulin antibody. Tubulin is detected with an apparent mass of 60 kDa. (C) The upper membranes represent two independent replicates of the experiment described in B. These membranes have been cut and incubated with anti-tubulin antibody. Ctrl, control noninfected neurons; MWM, molecular weight marker.
E46K Toxicity Correlates with Strong PLK2-Dependent S129 Phosphorylation, but Phosphorylation Does Not Explain E46K Toxicity.
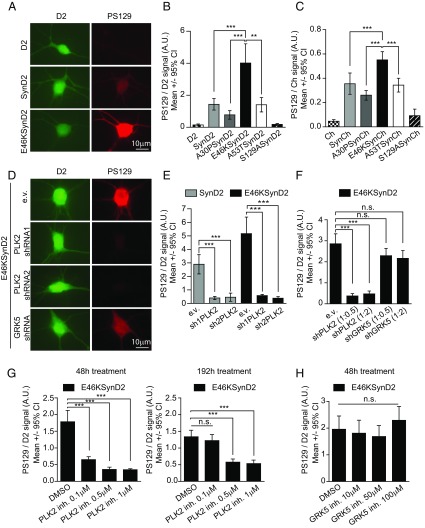
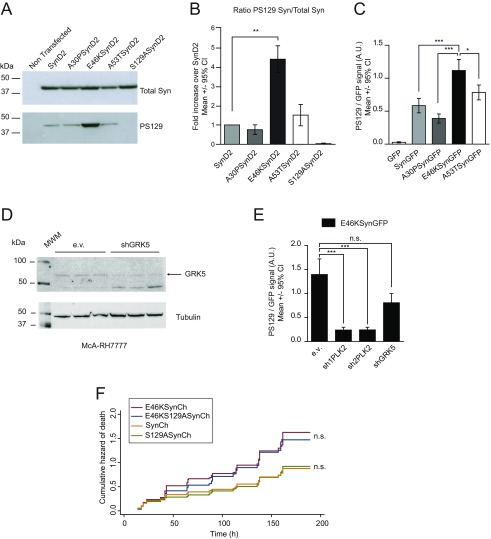
Posttranslational modifications such as S129 phosphorylation (PS129) could play a role in aSyn toxicity (14), and thus we measured the PS129 in neurons expressing the WT or mutant forms of aSyn by immunofluorescence using a PS129-specific antibody. The pathological mutation E46K exhibited the highest levels of PS129 (Fig. 4 A–C and Figs. S3C and S5 A–C), confirming recent data (43). Moreover, the extent of PS129 correlated with the degree of aSyn toxicity (Fig. 1B).
Fig. 4.
The E46K mutant exhibits the highest levels of PLK2-dependent PS129. (A) Immunofluorescence staining of primary rat cortical neurons expressing D2 or E46KSynD2 with a specific antibody against PS129. The green fluorescence signal from D2 is a surrogate for total aSyn levels, and the red fluorescence signal indicates the anti-PS129 binding (PS129 levels). (B) Quantification of the anti-PS129 binding over the D2 fluorescence intensity (total aSyn) in individual neurons expressing SynD2, A30PSynD2, E46KSynD2, or A53TSynD2; neurons expressing D2 and the S129ASynD2 mutant were used as negative controls (n = 20–40 neurons per condition; results of a representative experiment of three independent experiments are shown). (C) Similar results were obtained for aSyn Ch-tagged versions (n = around 30 neurons per condition). (D) Neurons cotransfected with E46KSynD2 and PLK2/GRK5 shRNAs containing plasmids or the empty vector (e.v.) as a control and immunostained with the anti-PS129 antibody. (E) Quantification of anti-PS129 binding relative to the D2 fluorescence intensity (total aSyn) in individual neurons expressing SynD2 or E46KSynD2 with two specific PLK2 shRNAs, sh1PLK2 and sh2PLK2. (n = 8–23 neurons per condition; results of a representative experiment of at least four independent experiments are shown). (F) Anti-PS129 binding relative to the D2 fluorescence intensity (total aSyn) in individual neurons expressing E46KSynD2 with PLK2- and GRK5-specific shRNAs at ratios of 1:2 and 1:0.5 (n = 30–44 neurons per condition from two independent experiments). (G) Quantification of PS129 in neurons expressing E46KSynD2 48 and 192 h after addition of the PLK2-specific inhibitor (inh). The effect persists at the higher doses 192 h after addition (n = 30–35 neurons per condition; results from a representative experiment of at least two independent experiments are shown). (H) A similar experiment using a GRK5-specific inhibitor did not decrease PS129 after 48 h (n = 20 neurons; results from a representative experiment of two independent experiments are shown). All error bars indicate 95% CIs; n.s., nonsignificant; **P < 0.01, ***P < 0.001 Kruskal–Wallis and Dunn’s post hoc test.
Fig. S5.
Analysis of PS129 of WT and mutant aSyn and its effect on neuronal survival. (A) HEK293 cells were transiently transfected with D2-tagged constructs of aSyn (WT, the pathological A30P, E46K, and A53T mutants, and the S129A mutant). The levels of PS129 were analyzed in Western blots using anti-Total aSyn and anti-PS129–specific antibodies. (B) Quantification of PS129 relative to the total aSyn levels in Western blots. The graph shows the values for each aSyn mutant normalized to WT aSyn (data are from three independent experiments; Kruskal–Wallis and Dunn’s post hoc test; **P < 0.01). (C) Quantification of PS129 binding over GFP intensity (total aSyn levels) in individual immunostained neurons expressing the WT and pathological aSyn GFP-tagged mutants SynGFP, A30PSynGFP, E46KSynGFP, and A53TSynGFP (data shown are from one representative experiment of three independent experiments; n = 20–39 neurons per condition; Kruskal–Wallis and Dunn’s post hoc test; *P < 0.05, ***P < 0.001). (D) Western blot of protein extracts from McA-RH7777 cells (rat cell line) transfected with a plasmid containing a specific GRK5 shRNA or the empty vector (e.v.) as a control. A specific antibody for GRK5 recognizes a band located at around 68 kDa, approximating the mass of GRK5. A decrease in GRK5 protein is observed in cells in which GRK5 shRNA is expressed. (E) Rat cortical primary neurons were cotransfected with plasmids containing E46KSynGFP and PLK2/GRK5 shRNAs or with the e.v. as a control (1:2 ratio) and were immunostained with anti-PS129 antibody. Quantification of anti-PS129 binding over GFP intensity (total aSyn levels) in individual neurons shows that only PLK2 shRNAs significantly decrease PS129 (n = 27–30 neurons per condition; Kruskal–Wallis and Dunn’s post hoc test; n.s., nonsignificant, ***P < 0.001). (F) Longitudinal survival of primary rat cortical neurons expressing Ch-tagged versions of aSyn (WT SynCh and the S129ASynCh, E46KSynCh, and E46KS129ASynCh mutants). The S129A mutant, which is PS129 incompetent, does not alter the risk of neuronal death (n = 500–650 neurons from two independent experiments; log-rank test; n.s., nonsignificant). All error bars represent 95% CIs.
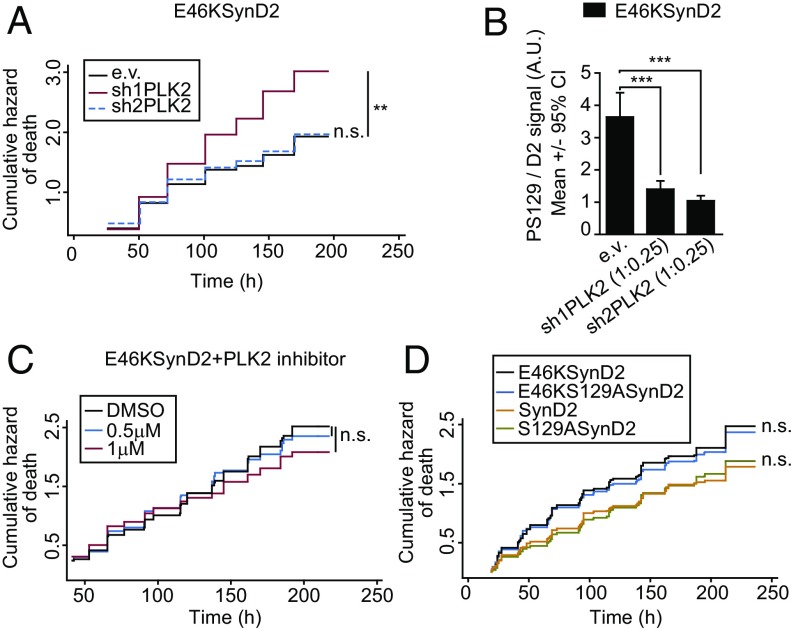
Two kinases, Polo-like kinase 2 (PLK2) and G protein-coupled receptor kinase 5 (GRK5), have been shown to phosphorylate S129 in neurons (44, 45). To assess the contribution of these two kinases to aSyn PS129 in our neuronal model, we cotransfected aSyn with plasmids encoding validated shRNAs against rat PLK2 (46, 47) or human/rat GRK5 (Fig. S5D) (48). Although both shRNAs seemed to reduce the PS129 of the WT and E46K aSyn, only PLK2 silencing yielded a significant inhibition (around 85% reduction) (Fig. 4 D–F and Fig. S5E). Indeed, a selective PLK2 inhibitor (49) inhibited PS129 in a dose-dependent manner, an effect that was observed 48 h after inhibition and that persisted for up to 8 d at the higher doses (Fig. 4G). By contrast, the GRK5 inhibitor amlexanox (50) did not diminish the PS129 aSyn (Fig. 4H), suggesting that PLK2 was the major driver of neuronal aSyn PS129.
Whether aSyn PS129 is causally related to neuronal death remains under debate. Given the close correlation between PS129 levels and aSyn-dependent toxicity, we tested whether PS129 contributes to neuronal death by inhibiting PLK2-dependent aSyn phosphorylation and by generating phosphorylation-incompetent aSyn mutants (S129A). In longitudinal survival analyses of primary cortical neurons cotransfected with E46K aSyn and PLK2 shRNAs, the PLK2 shRNAs decreased PS129, but they did not alter aSyn E46K-dependent toxicity (the increased toxicity with sh1PLK2 was probably due to off-target effects) (Fig. 5 A and B). As a complementary approach, we evaluated the effect of pharmacological inhibition of PLK2 on the survival of E46KSynD2–overexpressing neurons. Again, while the PLK2 inhibitor produced a decrease in aSyn PS129 (Fig. 4G), no changes in neuronal death were evident (Fig. 5C). Furthermore, longitudinal survival analysis of neurons expressing WT or mutant aSyn (E46K, S129A, or E46KS129A) demonstrated that blocking PS129 did not reduce E46K-dependent neuronal toxicity (Fig. 5D and Fig. S5F).
Fig. 5.
PLK2-dependent PS129 does not explain the E46K-dependent toxicity. (A) Cumulative hazard estimates of primary rat cortical neurons coexpressing E46KSynD2 and PLK2 shRNAs (sh1PLK2 and sh2PLK2) (log-rank test; n = around 150 neurons per condition). (B) Quantification of PS129 relative to the D2 fluorescence intensity (total aSyn) by immunostaining individual neurons expressing E46KSynD2 and PLK2 shRNAs with the specific anti-PS129 antibody. Under the same conditions used in A, PLK2 shRNAs significantly reduced E46KSynD2 PS129 (Kruskal–Wallis and Dunn’s post hoc test; n = 18–26 neurons per condition). (C) Neurons transiently transfected with E46KSynD2 were treated with two doses of the PLK2 inhibitor previously tested and subjected to longitudinal survival analysis (log-rank test; n = 600–1,000 neurons from five independent experiments). (D) Longitudinal survival of neurons expressing aSyn WT or the pathological E46K mutation ± the S129A mutation (CPH analysis; n = 2,500 neurons per condition from seven independent experiments). All error bars indicate 95% CIs; n.s., nonsignificant, **P < 0.01, ***P < 0.001.
Together, these results confirm PS129 to be a pathological hallmark of aSyn-dependent neurodegeneration and identify PLK2 as a major driver of this phosphorylation. However, as the turnover or aggregation of the heavily phosphorylated E46K protein was not altered in our experimental model, PLK2-dependent aSyn phosphorylation would appear to precede the formation of LBs, and as such it may represent an effective biomarker for early alterations in PD. Nevertheless, our results demonstrate that aSyn PS129 does not explain the toxicity of this protein, and they suggest that this pathological hallmark could be an epiphenomenon.
E46K Toxicity Is Predominantly Cell Autonomous.
Since our experimental approach documents the death of neurons overexpressing WT or mutant aSyn, and particularly the E46K mutation, it is tempting to assume a priori that toxicity occurs in a cell-autonomous manner. However, aSyn protein could propagate from neuron to neuron in distinct complexes (e.g., monomers, oligomers, or aggregates), spreading toxicity through a non–cell-autonomous mechanism. In addition to the mobilization of aSyn species from cell to cell, E46K toxicity could propagate to neighboring neurons by other means, such as through the release of cell debris after neuronal death or through changes in neuronal activity. For instance, overexpression of the E46K mutant in hippocampal neurons inhibits neurotransmitter release (51).
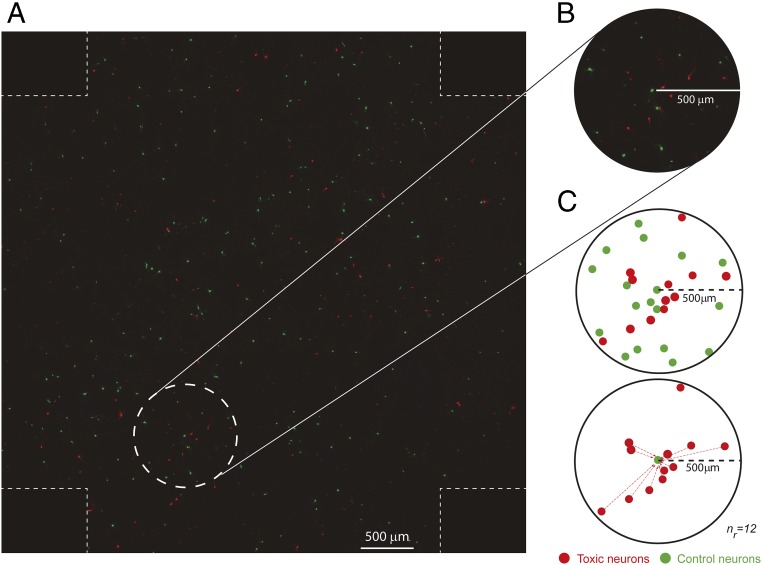
To establish the contribution of non–cell-autonomous mechanisms to aSyn toxicity, we developed an experimental set-up to measure whether neurons expressing a toxic protein affect their neighbors’ risk dying. Primary cortical neurons were transfected first with a plasmid expressing a control GFP protein and 24 h later with a second plasmid expressing the Ch-tagged toxic protein of interest. As the probability of cotransfected neurons is very low (around 0.2–0.3% of neurons), the resulting culture contains two different neuronal populations: control neurons expressing GFP (in green) and toxic neurons expressing Ch-tagged toxic proteins (in red) (Fig. 6A). This allows the risk of death of control neurons when surrounded by neurons expressing neurotoxic proteins to be scored. We acquired adjacent nonoverlapping fields of these neuronal cultures covering a total area of around 46 mm2 and combined them to create a single tiled image (Fig. 6A). “Green” and “red” neurons were tracked by automated microscopy for up to 8–9 d, and we estimated the individual survival time and assigned relative positions (x/y coordinates) for each individual neuron in the tiled image.
Fig. 6.
Experimental strategy to study the contribution of non–cell-autonomous mechanisms in neuronal death. (A) To assess the potential contribution of non–cell-autonomous mechanisms to neuronal death, we assessed how the risk of death in neurons expressing a control protein (i.e., GFP) is influenced by neighboring neurons expressing a toxic protein (i.e., Ch-tagged aSyn constructs). To that goal, we longitudinally imaged primary rat cortical neurons transiently transfected with control and toxic proteins in the same field. Shown is an example of a single tiled image of these neurons created with adjacent nonoverlapping images. Individual survival times were estimated for each individual neuron, and a CPH analysis was used to analyze how toxic neurons (red) influenced the risk of death of control neurons (green). (B) Magnified image of the area within the dashed circle in A showing toxic and control neurons within a 500-μm radius. (C) The number of toxic and control neurons within a 500-μm radius around a single control neuron. The number of red (toxic) neurons in a particular radius around each single GFP (control) neuron is the variable nr (in the example, nr = 12). The nr was calculated for each single GFP neuron in the tiled image at radii of 500 μm and 1,000 μm.
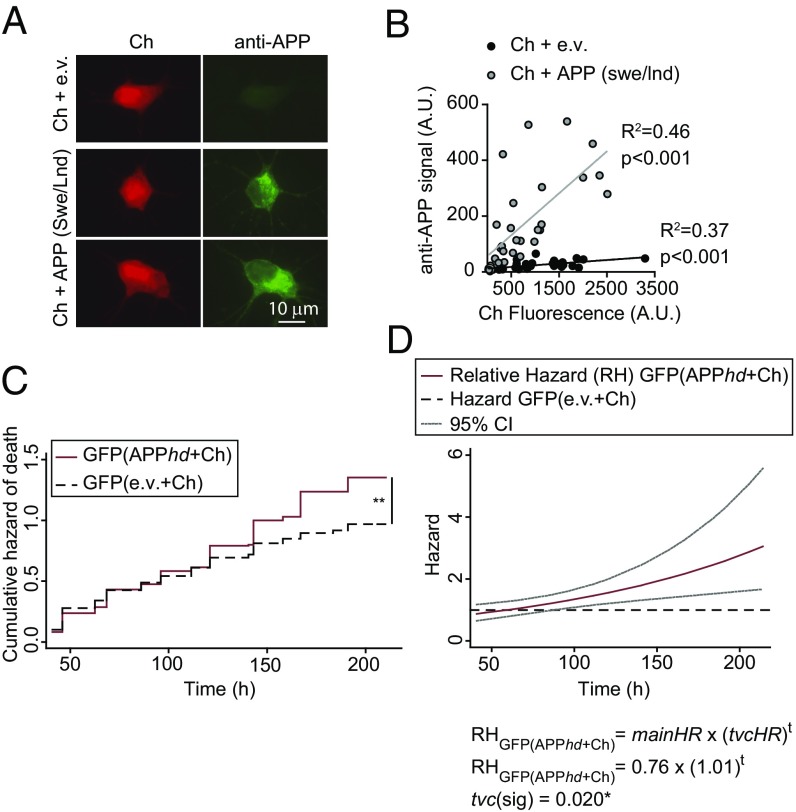
To validate the approach, we overexpressed an established inducer of nonautonomous toxicity, mutant human amyloid precursor protein (APP). In Alzheimer disease (AD), abnormal APP leads to the accumulation of amyloid-β (Aβ) extracellular aggregates that impair synaptic function and trigger neuronal loss (52). Pathological APP mutations increase both the amount and aggregation propensity of Aβ peptides. Extracellular Aβ secreted by neurons expressing APP harboring the Swedish and London mutations (APPswe/lnd) impairs long-term potentiation of adjacent neurons that do not express APPswe/lnd (53). We transfected primary cortical neurons with GFP (as a control) followed by cotransfection with constructs expressing the APPswe/lnd mutant (53) and Ch (APP+Ch). Taking advantage of the strong correlation between APP and Ch expression (Fig. 7 A and B), we used red fluorescence as a surrogate for APP expression. The risk of death for APP+Ch-expressing neurons was estimated by a CPH analysis, and, as expected, neurons expressing APPswe/lnd had a high risk of death, increasing at higher doses (hd) of the APP plasmid (APP+Ch HR, 1.65; APPhd+Ch HR, 2.46) (Table 1). We then scored the risk of death of control GFP neurons neighboring APP+Ch neurons; higher doses of APP significantly affected the survival of control cells [GFP(APPhd+Ch) HR 1.31] (Table 1). The risk of death increased during the experiment, such that a significant risk of death could only be detected at the later time points (Fig. 7C). Hence, a CPH analysis was performed considering the variable group as a time-varying covariate (tvc), which confirmed that the relative hazard of GFP neurons cocultured with (APPhd+Ch)-expressing neurons changed over time, following the equation: HR = 0.76 × (1.01)t (Table S2). The effect of (APPhd+Ch)-expressing neurons over GFP control neurons starts to be significant 100 h after transfection (Fig. 7D). Therefore, our experimental set-up is sensitive to detect non–cell-autonomous toxicity in neurons.
Fig. 7.
Neurons expressing APPswe/Lnd increase the risk of death of neighboring control neurons. (A) Primary rat cortical neurons were cotransfected with plasmids expressing APPswe/lnd or the empty vector (e.v.) and Ch and were immunostained with an APP-specific antibody. (B) Fluorescence intensity from Ch and from secondary antibodies recognizing APP was quantified in individual neurons. A correlation analysis was performed showing that Ch+ neurons expressed the APPswe/lnd protein compared with control conditions (e.v.). (C) Cumulative risk estimates of GFP-expressing neurons neighboring APP-expressing neurons transfected with a high dose of APP+Ch (APPhd+Ch) or control neurons (e.v.+Ch). CPH analysis of 250–300 neurons from two independent experiments: **P < 0.01. (D) Cox modeling of the time-dependent variation in the relative hazard in a population of neighboring APP-expressing neurons [GFP(APPhd+Ch)] with respect to a population of neighboring control neurons [GFP(e.v.+Ch)]. The main relative hazards at t = 0 and tvc are from Table S2 95% CIs.
Table 1.
CPH model of the effect of APPswe/lnd on neuronal survival
| Group | HR | 95% CI | P value |
| (APP+Ch) (n = 838) vs. (e.v.+Ch) (n = 812) | 1.65 | 1.11−2.46 | 0.014* |
| (APPhd+Ch) (n = 643) vs. (e.v.+Ch) (n = 703) | 2.46 | 2.02−2.99 | <0.001*** |
| GFP(APP+Ch) (n = 515) vs. GFP(e.v+Ch) (n = 471) | 1.10 | 0.91−1.33 | 0.310 |
| GFP(APPhd+Ch) (n = 294) vs. GFP(e.v.+Ch) (n = 254) | 1.31† | 1.08−1.58 | 0.006** |
n, number of neurons. Data are from three independent experiments.
Violates proportional hazard assumption (P = 0.013*).
Table S2.
Time-dependent effect of APPhdswe/lnd on neuronal survival
| Group | HR | 95% CI | P value |
| Main: GFP(APPhd+Ch) | 0.76 | 0.50–1.15 | 0.196 |
| Tvc: GFP(APPhd+Ch) | 1.01 | 1.00–1.01 | 0.020* |
Main, relative hazard at t = 0. Data are from two independent experiments.
We next evaluated whether neurons overexpressing Ch-tagged WT and E46K aSyn could influence the survival of neighboring GFP-expressing neurons. WT and E46K aSyn overexpression significantly increased the risk of neuronal death (SynCh HR = 1.33 and E46KSynCh HR = 1.94) (Table 2). However, neither GFP neurons that neighbored SynCh-expressing neurons nor GFP neurons neighboring E46KSynCh-expressing neurons experienced a significant increase in the risk of death compared with control conditions (GFP neurons neighboring Ch-expressing neurons) (Table 2).
Table 2.
CPH model of the effect of aSyn (WT and mutant E46K) on neuronal survival
| Group | HR | 95% CI | P value |
| SynCh (n = 372) vs. Ch (n = 390) | 1.33 | 1.08−1.64 | 0.008** |
| E46KSynCh (n = 371) vs. Ch (n = 390) | 1.94 | 1.71−2.19 | <0.001*** |
| GFP(SynCh) (n = 581) vs. GFP(Ch) (n = 684) | 1.03 | 0.73−1.44 | 0.883 |
| GFP(E46KSynCh) (n = 823) vs. GFP(Ch) (n = 684) | 0.96 | 0.65−1.42 | 0.851 |
n, number of neurons. Data are from two independent experiments.
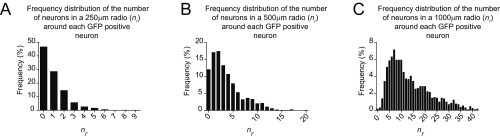
The previous experiments analyzed the nonautonomous toxicity in a manner independent of distance (i.e., the effect comes from the whole set of neurons within the tiled image). Nevertheless, nonautonomous toxicity could be further modeled as a function of the number of neurons that are located in close vicinity and express a neurotoxic factor. If that were the case, the proximity and number of neurons expressing a neurotoxic protein would affect the risk of death of control neurons. Taking this possibility into account, we defined the variable, nr, that accounts for the number of neurons expressing a neurotoxic protein (SynCh or E46KSynCh), or Ch as a control, within a circle of radius (r) centered on each GFP+ control neuron (Fig. 6 B and C). The radii of r = 500 μm and r = 1,000 μm were investigated based on the frequency distribution of nr for the whole set of GFP neurons in the tiled image (Fig. S6). Subsequently, we estimated the risk of death of GFP+ neurons by using CPH models adjusted to nr (Table 3 and Table S3). In the adjusted model (with the group and nr factors), the group factor (SynCh or E46KSynCh) did not show any effect on the risk of death of neighboring GFP+ neurons (compared with neighboring Ch neurons). On the other hand, nr did have a significant influence on this parameter (Table 3). Interestingly, a significant interaction between the group and nr factors was evident for both r = 500 μm and r = 1,000 μm (Wald test, P < 0.001). In particular, we found a small but significant effect on the risk of death of GFP neurons neighboring E46KSynCh neurons (HR 1.03 for r = 500 μm and HR 1.01 for r = 1,000 μm) but not for the GFP neurons neighboring SynCh neurons (Table 3 and Table S3). Hence, this mild nonautonomous effect observed in our E46KSynCh primary cultures appeared to decrease over distance and escapes the detection threshold at the whole-population level. Of note, further microscopy analysis gave no evidence of cell-to-cell spread of E46K aSyn species to GFP control neurons (Fig. S7) that could explain this mild nonautonomous effect.
Fig. S6.
Graphs showing the frequency (%) distribution of red neurons expressing Ch, SynCh, and E46KSynCh in a fixed radius (250 μm, 500 μm, or 1,000 μm) (nr) around each GFP+ neuron. (A) 250-μm radius. (B) 500-μm radius. (C) 1,000-μm radius. In radii lower than 500 μm, the frequency of GFP neurons without neurons around them is very high (around 46% of total neurons).
Table 3.
CPH model of the effect of nr (500-μm radius) on neuronal survival
| Variable | HR | 95% CI | P value |
| Group | |||
| GFP(SynCh) (n = 501) vs. GFP(Ch) (n = 590) | 1.09 | 0.70–1.70 | 0.694 |
| GFP(E46KSynCh) (n = 645) vs. GFP(Ch) (n = 590) | 0.90 | 0.57–1.43 | 0.662 |
| nr | 0.96 | 0.94–0.99 | 0.005** |
| Interaction group # nr | |||
| GFP(SynCh) (n = 501) vs. GFP(Ch) (n = 590) | 0.99 | 0.94–1.03 | 0.576 |
| GFP(E46KSynCh) (n = 645) vs. GFP(Ch) (n = 590) | 1.03 | 1.00–1.06 | 0.034* |
n, number of neurons; nr, number of red neurons (Ch, SynCh, E46KSynCh) in a 500-μm radius around each single GFP+ neuron. #, interaction. Data are from two independent experiments.
Table S3.
CPH model of the effect of nr (1,000-μm radius) on neuronal survival
| Variable | HR | 95% CI | P value |
| Group | |||
| GFP(SynCh) (n = 402) vs. GFP(Ch) (n = 473) | 1.03 | 0.60−1.75 | 0.927 |
| GFP(E46KSynCh) (n = 443) vs. GFP(Ch) (n = 473) | 0.87 | 0.52−1.46 | 0.592 |
| nr | 0.98 | 0.97−0.99 | <0.001*** |
| Interaction group # nr | |||
| GFP(SynCh) (n = 402) vs. GFP(Ch) (n = 473) | 1.00 | 0.98−1.02 | 0.947 |
| GFP(E46KSynCh) (n = 443) vs. GFP(Ch) (n = 473) | 1.01 | 1.00−1.02 | 0.019* |
nr, number of red neurons (Ch, SynCh, E46KSynCh) in a 1,000-μm radius around each single GFP+ neuron. #, interaction. Data are from two independent experiments.
Fig. S7.
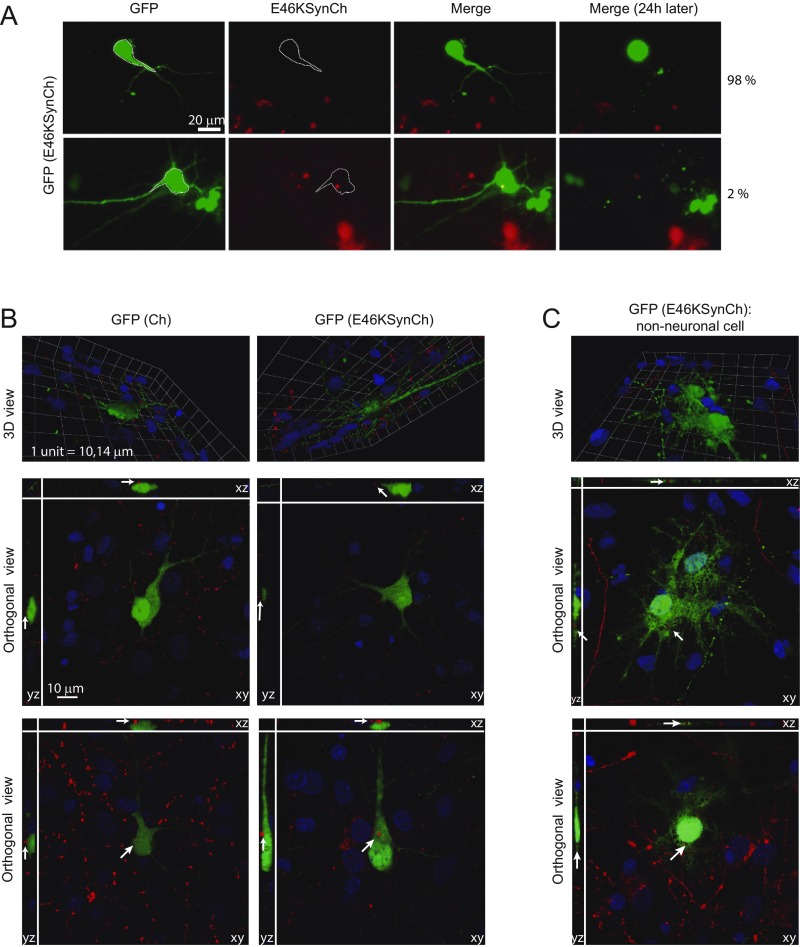
Evaluation of cell-to-cell aSyn spread in dying GFP control transfected cells neighboring E46KSynCh-expressing neurons. GFP living neurons that were dead in the subsequent 24 h were considered as “dying neurons.” Images from our longitudinal survival experiments enabled us to select dying GFP control neurons neighboring E46KSynCh neurons and to analyze whether they had red signal inside as a surrogate for potential uptake of aSyn species. (A) From a total of 100 dying control neurons analyzed, we found only two neurons with red signal apparently inside. (B) Confocal analysis of GFP-transfected cells neighboring Ch- or E46KSynCh-expressing neurons. Neurons were fixed at 8 dpt. (Top Row) Side view of 3D reconstructed images of GFP-transfected neurons with Ch-expressing neighboring neurons (Left) and with E46KSynCh-expressing neighboring neurons (Right). (Middle and Bottom Rows) Orthogonal views from different planes (x/y, x/z, and y/z) of confocal microscope images. Nuclei were stained with DAPI (blue); the green signal is GFP, and the red signal corresponds to Ch or E46KSynCh. Shown are representative images of GFP-expressing neurons neighboring Ch-expressing neurons (n = 30) or E46KSynCh-expressing neurons (n = 51). Red particles were not found inside GFP-expressing neurons. Similar results were obtained in neurons fixed at 4 dpt. (C) Confocal analysis of GFP-transfected nonneuronal cells neighboring E46KSynCh-expressing neurons. (Top) Side view of 3D reconstructed images from the GFP-transfected cell with E46KSynCh-expressing neighboring neurons. (Middle and Bottom) Orthogonal views from different planes (x/y, x/z, and y/z) of confocal microscope images. Nuclei were stained with DAPI (blue); the green signal represents GFP, and the red signal corresponds to Ch or E46KSynCh. Red particles were found inside GFP-expressing nonneuronal cells.
Therefore, and although we were able to identify a minor nonautonomous component that was spatially restricted, E46K aSyn toxicity is predominantly cell autonomous.
Discussion
We describe here an in vitro neuronal model that recapitulates aSyn toxicity based on the expression of fluorescently tagged aSyn in primary cultures of rat cortical neurons (WT and the pathological A30P, E46K, and A53T mutants), coupled to the longitudinal tracking of individual neurons over a protracted time course by automated microscopy. Our experimental set-up enables a longitudinal survival analysis to be applied, yielding quantitative estimations of the risk of neuronal death associated with aSyn and allowing its toxicity to be investigated. Through this approach, we were able to identify the pathological E46K aSyn mutation as the most toxic variant, although its enhanced toxicity was not due to enhanced protein stability or aggregation, and it was triggered by soluble aSyn species. We also found that a main pathological feature of PD, S129 aSyn phosphorylation, was strongly correlated with aSyn toxicity, such that the strongest PS129 was associated with the E46K mutation. PLK2 was identified as the main S129 kinase in neurons, but independent approaches demonstrated that PS129 does not play a causative role in neurotoxicity and that, instead, it probably represents an epiphenomenon. Nevertheless, the strong correlation between PS129 and the risk of neuronal death suggests that it could be a useful biomarker for early disease stages. Finally, we evaluated whether E46K-expressing neurons confer susceptibility to death to neighboring control neurons. While we identified a minor E46K-dependent nonautonomous toxic component, it was spatially restricted to local neighbors. Thus, in our experimental set-up E46K aSyn toxicity is predominantly due to cell-autonomous mechanisms.
Patients with mutations in the SNCA gene often display cognitive impairment (54), which appears to be strongly associated with the E46K mutation. Indeed, the patients in which this mutation was first described developed severe cognitive decline, dementia, and visual hallucinations, all features associated with DLB (55). This is consistent with our results showing E46K to be the most toxic mutation in cortical neurons. We also found that the A53T mutation decreases neuronal survival, albeit to a lesser extent than the E46K mutation. Notably, the toxicity associated with WT aSyn reached statistical significance only with Ch-tagged versions, probably because of the low intrinsic toxicity that this fluorescent protein displays. Toxicity of D2-tagged WT aSyn was reported recently, but differences in the expression vector used or the fact that fusions were performed N-terminally could account for this discrepancy (35).
The toxicity evident in our in vitro system (E46K > A53T > WT) has been observed in mouse models in which aSyn variants are overexpressed in the substantia nigra by lentiviral transduction (41). Intriguingly, our results did not recapitulate the A30P-associated toxicity, a mutant that stands apart from the other mutants in different experimental assays. For instance, A30P aSyn has the slowest propensity to induce fibril formation in vitro (56), and it is equally or less toxic than WT aSyn in yeast models (57). It is noteworthy that the A30P mutation disrupts the membrane association of aSyn (58). Indeed, overexpression of WT, E46K, and A53T aSyn in hippocampal primary neurons inhibits synaptic vesicle exocytosis and neurotransmitter release, while the A30P mutant does not (51). Given these differences, it is reasonable to assume that the mechanism of A30P toxicity could differ from that of the other aSyn mutants.
Compared with animal models of aSyn toxicity, the primary culture model described here recapitulates neuronal loss. Overexpression of human E46K aSyn in rodents reproduces motor impairment and the formation of aSyn intraneuronal aggregates resembling LBs, but neuronal death was not reported (59, 60). Similarly, most WT and mutant aSyn transgenic mouse models reproduce features of PD, such as striatal dopamine loss, aSyn aggregation, and/or motor impairment, but not neuronal death in the substantia nigra (61). Nevertheless, neuronal death has been reported in a conditional murine model expressing WT and A30P aSyn and in a BAC model expressing WT aSyn (62–64). One major advantage of our model is that it recapitulates aSyn-dependent toxicity in a short temporal framework of 8–9 d, and toxicity can be scored quantitatively to explore the molecular mechanisms of aSyn toxicity as well as therapeutic approaches aimed at increasing the survival of neurons.
It has been reported that the pathological A30P and A53T mutations interfere with chaperon-mediated autophagy, thereby altering the degradation of aSyn and other substrates (12). Since increased aSyn steady-state levels are sufficient to promote PD, we determined whether the E46K mutation altered the aSyn half-life in primary living neurons using OPL as an indirect assessment of altered degradation. E46K aSyn turnover was similar to that of the WT protein and was even higher than that of A30P aSyn, ruling out altered protein turnover as a driver of E46K toxicity.
Both WT and mutant aSyn concentrate into insoluble oligomers or aggregates of different sizes within neurons, and these species are commonly found in patients and disease models. However, we did not detect overt aggregation in the time course of our experiments, suggesting that protein aggregation is not required for aSyn neurotoxicity. In agreement with this idea, analysis of postmortem human brains affected by synucleinopathies unveiled a dissociation between LBs and neuronal death (65–67). Similar dissociation between aggregation and neuronal death has been reported in experimental models of other neurotoxic proteins [i.e., mutant huntingtin or leucine-rich repeat kinase 2 (LRRK2)] (30, 35) as well as aSyn (64, 68). Thus, our study further supports the notion that aSyn toxicity may arise from early intermediates in the aggregation process (69) or that it is independent of aggregation.
Importantly, we observed differences in aSyn phosphorylation, a pathological hallmark of synucleinopathies, with E46K being the most strongly phosphorylated form, in agreement with recent studies (43). Using specific shRNAs and kinase inhibitors, we identified PLK2 as the kinase preferentially involved in this phosphorylation. PLK2 phosphorylates aSyn S129 in the CNS (44), and it is more concentrated in dopaminergic neurons of aged monkeys (70). The pathological meaning of this posttranslational modification remains unclear. PS129 has been proposed to increase aSyn fibrillization (71) and favor autophagy-dependent aSyn degradation (72), but controversial results have been obtained when trying to link PS129 to neuronal death (15). Our data indicate that PS129 occurs in the absence of aggregation and that it does not alter aSyn turnover (which is even increased in E46K relative to other mutants such as A30P). Here we found a striking correlation between aSyn PS129 and neuronal toxicity, and since PLK2 seems to be the main kinase involved in this modification, we could directly test the contribution of PS129 to neuronal death. Through three independent approaches (modulating PLK2 levels and activity and preventing PS129 with the mutation S129A), we demonstrate that impeding aSyn PS129 does not inhibit neuronal death, indicating that this pathological hallmark is an epiphenomenon. We can hypothesize a scenario in which an increase in PLK2-dependent PS129 coincides with the establishment of toxicity without necessarily contributing to neuronal death. PLK2 is an activity-dependent kinase, the levels of which increase with elevated synaptic activity. In this scenario, PLK2 might phosphorylate substrates involved in rearrangements of the actin cytoskeleton, favoring the spine remodeling that would down-regulate synaptic activity (46, 73). If enhanced aSyn expression leads to changes in neuronal activity, PLK2 could be activated and increase PS129 as part of a program to promote specific substrate degradation and restore neuronal activity. Further experiments should be performed to test this hypothesis.
A growing body of evidence suggests that aSyn species can be released to the extracellular milieu and translocate into neighboring cells by endocytosis. Although the precise mechanism of transmission is not well understood, the hypothesis that aSyn spreading accounts for nonautonomous neuronal death in PD is currently under debate. While experimental evidence demonstrates that aSyn can promote neuronal death via cell-autonomous and nonautonomous mechanisms, the relative contribution of both mechanisms to toxicity has yet to be established. In our system, E46K aSyn-expressing neurons only marginally influenced the survival of neighboring control neurons, such that only control neurons near E46K aSyn neurons were affected. This effect was largely dependent on the distance and number of toxic neighbors, failing to detect a nonautonomous effect in the whole-population analysis. Although cell-to-cell spread of E46K aSyn could be a tempting explanation for this mild nonautonomous effect, we did not find evidence supporting this notion. By contrast, and as expected, nonautonomous toxicity was detected when APP was overexpressed. This toxicity was time dependent, suggesting that Aβ peptides accumulate in the culture over time. We cannot rule out the possibility that under more toxic regimes, or over longer time scales, nonautonomous E46K toxicity may play a more relevant role. Nevertheless, our results clearly illustrate a more important cell-autonomous component.
Taken together, our model of aSyn toxicity revealed that some of the main hallmarks of aSyn pathology are not necessary to drive neuronal death. We postulate that the cell-autonomous toxicity arising from nonaggregated forms of E46K may recapitulate early events in aSyn toxicity. Among them, PLK2-dependent aSyn phosphorylation could represent an early biomarker of aSyn toxicity that precedes protein aggregation, while the spreading of aSyn through neurons may preferentially occur at later stages of the disease, in the presence of oligomeric species or protein aggregates. Among the aSyn toxic mechanisms proposed, alterations to vesicle trafficking and neurotransmission could underlie E46K toxicity, as well as interference with the endoplasmic reticulum–Golgi transport and the induction of the unfolded protein response (74). Finally, this aSyn neurotoxicity model could serve not only to explain early pathological events of synucleinopathies but also could constitute a platform to assess if new therapeutic approaches enhance the survival of neurons affected by these pathologies.
Materials and Methods
Plasmids.
Detailed information on the plasmids used in this study is provided in SI Materials and Methods. All plasmids were verified by DNA sequencing.
Neuronal and Cell Cultures, Transfections, and Lentiviral Infections.
Animal handling was carried out in accordance with European Community Council Directive 2010/63/EC and Spanish legislation (Real Decreto 53/2013), and the protocols were all approved by the Ethics Committee of the University of Navarra (051-13). Primary cortical neurons were obtained from E20 Sprague–Dawley rat embryos (Charles-River Laboratories). Neurons, HEK293 cells, and McA-RH7777 cells were transfected with Lipofectamine 2000 (Invitrogen) or calcium phosphate after 5 d in vitro. For further details see SI Materials and Methods.
Sequential Biochemical Fractionation to Detect Insoluble aSyn Species.
Sequential biochemical fractionation of whole-cell lysates from primary cortical neurons (modified from ref. 75) was performed to separate 1% Tx-100–soluble and –insoluble aSyn species. For further details see SI Materials and Methods.
Protein Extraction, SDS/PAGE, and Western Blotting.
Whole-cell lysates were collected in radioimmunoprecipitation assay (RIPA) buffer 2 d posttransfection (dpt). After protein quantification, the samples were separated by SDS/PAGE and were transferred to nitrocellulose membranes (Bio-Rad Laboratories). The membranes were blocked and then were probed with primary antibodies overnight at 4 °C and with secondary antibodies for 2 h at room temperature (for further details see SI Materials and Methods).
Drug Treatments.
To evaluate the contribution of the two different kinases to aSyn phosphorylation, two specific kinase inhibitors were applied to the neuronal cultures. Neurons were fixed with a 4% paraformaldehyde (PFA) (Panreac), 4% sucrose (Sigma) solution and visualized by immunofluorescence (for further details see SI Materials and Methods).
Immunofluorescence.
After fixation with PFA and 4% sucrose, neurons were permeabilized in 0.1% Tx-100, blocked with 3% goat serum (Jackson Immunoresearch) and 3% BSA (Merck-Millipore), and incubated with primary and secondary antibodies for 2 h at room temperature. The coverslips were finally stained with DAPI (Sigma) and observed at 63× magnification on a Zeiss Axiovert 200M fluorescence microscope (for further details see SI Materials and Methods).
Automated Image Acquisition.
Neuronal survival was studied by automatic longitudinal tracking of neuronal cultures every 12–24 h after transfection on a Zeiss Observer Z1 microscope (29) (for further details see SI Materials and Methods).
Image Processing and Statistics.
Fluorescence intensity was measured with MetaMorph Analysis software (Molecular Devices), and graphs were generated with GraphPad Prism 5 software. Survival and photoswitching images were analyzed with MATLAB-based custom programs, and survival analysis was performed with STATA 12 (for further details see SI Materials and Methods).
Antibodies.
Detailed information on the antibodies used in this study is available in SI Materials and Methods.
SI Materials and Methods
Plasmids.
D2 was derived from pDendra2-N (Evrogen) and subcloned into pCAGGs empty vector (a generous gift from R. Edwards, University of California, San Francisco). SynD2 fusions (WT/A30P/A53T) were derived from GFP–α-synuclein, GFP–α-synuclein (A53T), and GFP–α-synuclein (A30P) [a generous gift from R. Edwards (58)] by PCR amplification followed by subcloning into pCAGGs-D2. The E46KSynD2 construct was generated from GFP–α-synuclein by three-step PCR amplification using specific oligonucleotides to introduce the E46K mutation (synE46K forward: GGCTCCAAAACCAAGAAGGGAGTGGTGCATGG, and synE46K reverse: CCATGCACCACTCCCTTCTTGGTTTTGGAGCC), followed by subcloning into pCAGGs-D2. S129ASynD2 and E46KS129ASynD2 were generated from GFP–α-synuclein and E46KSynD2, respectively, by three-step PCR amplification using specific oligonucleotides to introduce the S129A mutation (synS129A forward: GGCTTATGAAATGCCTGCTGAGGAAGGGTATCAAG, and synS129A reverse: CTTGATACCCTTCCTCAGCAGGCATTTCATAAGCC), followed by subcloning into pCAGGs-D2. The pCAGGs-GFP plasmid was a generous gift from S. Finkbeiner, University of California, San Francisco. PCR amplification of GFP from pEGFP-N1 (Clontech) substituted D2 to generate the SynGFP fusions in pCAGGs vector. Ch was derived from SOD1-mCherry by PCR amplification (a generous gift from T. Aragón, University of Navarra, Pamplona, Navarra, Spain) and was subcloned into the pCAGGs vector. PCR amplification of Ch from pTAP242 (T. Aragón), and substitution of D2 generated the SynCh fusions (WT/A30P/E46K/A53T/) in the pCAGGs vector. The S129A Ch-tagged mutants were derived from the SynD2 fusions (S129A/E46KS129A) subcloned into pCAGGs-mCherry. PLK2 was derived from pCMV6-XL5 PLK2 (OriGene) and was subcloned into the pCAGGs vector. APPswe/lnd was derived from pCITE(APPswe/lnd) [a generous gift from S. Knafo, University of the Basque Country–Spanish National Research Council, Leioa, Bizkaia, Spain (53)], and it was subcloned into the pCAGGs vector. For the generation of pCAGGs-IRES-GFP, GFP derived from peGFP-N2 was subcloned into pIRES (a generous gift from R. Hernández, University of Navarra, Pamplona, Navarra, Spain) to generate pIRES-GFP. The IRES-GFP sequence was derived from pIRES-GFP and was subcloned into pCAGGs. Generation of pCAGGs-Syn (WT, E46K)-IRES-GFP WT and mutant E46K aSyn versions were derived by PCR amplification from pCAGGs-Syn (WT, E46K)-D2 plasmids with specific oligonucleotides introducing a stop codon after the aSyn coding sequence (Oligo forward, NheI: GCTCAAGCTAGCCCATGGATGTATTCAT, Oligo reverse stop, MluI: GCACGCGTTTAGGCTTCAGGTTCGTAGTCTTGA) and were subcloned in pCAGGs-IRES-GFP. pSuper-mCherry was derived from pSuper.gfp/neo (pSuper RNAi System from OligoEngine) (a generous gift from J. Wesseling, Instituto de Neurociencias–Spanish National Research Council, San Juan de Alicante, Alicante, Spain). To target selected sequences, the following oligonucleotides were introduced in pSuper-mCherry:
Neuronal and Cell Cultures.
Primary cultures of cortical neurons were established from E20 Sprague–Dawley rat embryos (Charles-River Laboratories) as described elsewhere (30), with minor modifications. Pregnant rats were killed with CO2, and their embryos were extracted and decapitated. The brain cortices were isolated from the embryos and dissociated with 1.5% papain (Worthington) for 10 min at 37 °C, followed by a 10-min treatment with DnaseI (Sigma). Finally, the tissue was treated for 20 min with a trypsin inhibitor (Sigma) to inhibit the papain, and individual neurons were obtained by mechanical trituration in Opti-MEM (Gibco) supplemented with 0.8% glucose (Sigma), gentamicin (Gibco), and fungizone (Gibco). The neurons were plated at a density of 5 × 105 neurons per well in 24-well plates (Costar) coated with laminin (BD Biosciences) and poly-d-lysine (Millipore), prewarmed to 37 °C. For immunofluorescence, 2.5 × 105 neurons per well were plated in 24-well plates with glass coverslips (Thermo Scientific) coated as described previously. After 1 h, once the neurons had attached to the coverslips, the Opti-MEM was replaced with Neurobasal medium (Gibco) supplemented with 5% FBS (HyClone), 1% GlutaMAX (Gibco), 2% B27 (Gibco), gentamicin, and fungizone. The neurons were then incubated at 37 °C in 5% CO2 for 5 d until transfection or lentiviral infection.
HEK293 or McA-RH7777 cells were maintained in 5% CO2 at 37 °C in 75-cm2 flasks (Corning) in 15 mL of DMEM (Gibco) supplemented with 10% FBS (Gibco), 1% sodium pyruvate (Gibco), and 1% penicillin-streptomycin (Gibco). Confluent cells were detached with Trypsin-EDTA (Sigma), and 20% of them were seeded in a new flask for maintenance or in six-well plates for transfection.
Calcium Phosphate Transfection of HEK293 Cells.
HEK293 cells were transfected using calcium phosphate. The cells were plated in six-well plates as described above on the day before transfection to reach an optimal 50% confluence on the day of transfection. One hour before the DNA-containing calcium phosphate precipitate was added, the culture medium was replaced with DMEM without FBS. The cells were transfected with the desired plasmid (4 μg) using two to three wells per condition. For the cotransfection of two different plasmids, 3 μg of each plasmid were used per well, and a total of 100 μL of the precipitate solution was added. The cells were incubated for 2–5 h at 37 °C, and once the calcium phosphate precipitate was visible as a thin granulate in the culture, the medium was replaced with 2 mL of DMEM plus 10% FBS per well.
Lipofectamine Transfection of Primary Rat Cortical Neurons and McA-RH7777 Cells.
Neurons or McA-RH7777 cells were transfected with the Lipofectamine 2000 transfection reagent (Invitrogen) after 5 d in vitro. One hour before the DNA mixture was added, the culture medium was replaced with Neurobasal or DMEM for neurons or McA-RH7777 cells, respectively. For lipofectamine complex formation, DNA and the lipofectamine reagent were diluted in Opti-MEM medium and were mixed in equal volumes. After incubation at RT for 20 min, 50 μL or 200 μL of the mixture was added to the wells of 24-well or six-well plates, respectively. Plates were replaced in the incubator, and 2–3 h later, the cells were washed and the medium was replaced with Neurobasal 1% FBS, 1% GlutaMAX (Gibco), 2% B27 (Gibco), gentamicin, and fungizone in the case of neurons or with DMEM with 10% FBS, 1% sodium pyruvate, and 1% penicillin-streptomycin in the case of McA-RH7777 cells.
Lentiviral Infection of Primary Rat Cortical Neurons.
Plasmids for lentiviral expression of aSyn (L3012 IRES-driven GFP reporter containing WT or E46K aSyn) were a generous gift from T. Südhof, Stanford University School of Medicine, Stanford, CA (41). With these plasmids, a set of lentiviruses carrying GFP, WT, or E46K aSyn (8.22 × 107, 2.28 × 108, and 2.18 × 108 infective units/mL, respectively) were generated by VIVEbioTECH. Cortical primary neurons were plated on six-well plates at a density of 2 × 106 neurons per well in Neurobasal medium + 5% FBS. After 5 d the medium was replaced with Neurobasal medium containing 1% FBS, and 10 μL of the lentiviral solution was added to three wells per condition (20 × 105 infective units per well of six-well plates). The neurons were maintained in a separate incubator until protein extraction.
Sequential Biochemical Fractionation for the Detection of Insoluble aSyn Species.
The aSyn species insoluble in 1% Tx-100 were considered aggregate forms. The neurons were washed 5 and 10 d after infection and then were recovered in ice-cold PBS (Lonza). Samples were centrifuged at 9,300 × g, and the pellets were resuspended in Tx-100 [1% Tx-100 (Sigma), 150 mM NaCl (Sigma), 50 mM Tris (Sigma) (pH 7.4), 5 mM EDTA (Sigma), 2 mM Na3VO4 (Sigma), 25 mM NaF (Sigma), and a protease inhibitor mixture (1 tablet/10 mL) (Roche)]. After sonication and incubation for 20 min on ice, extracts were centrifuged at 100,000 × g at 4 °C for 30 min. Supernatants were stored as the Triton-soluble fraction, and the pellets were resuspended in SDS buffer [2% SDS (Sigma), 150 mM NaCl, 50 mM Tris (pH 7.4), 5 mM EDTA, 2 mM Na3VO4, 25 mM NaF, and a protease inhibitor mixture (1 tablet/10 mL) (Roche)] to solubilize the 1% Tx-100–resistant species. These samples were then centrifuged at 16,000 × g at 25 °C for 10 min, and the supernatants were collected as the Triton-insoluble fraction.
Protein Extraction.
Two days posttransfection, whole-cell lysates from confluent HEK293 or McA-RH7777 cells were collected for protein extraction and Western blotting. The whole process was performed on ice. The culture medium was removed, and the cells were washed and scraped into 1 mL of cold PBS per well. The cells were centrifuged at 9,300 × g and 4 °C for 1 min, and the pellets were resuspended in 50 μL of RIPA-buffer per well [150 mM NaCl, 50 mM Tris (pH 7.5), 1% Tx-100, 0.1% SDS, 0.5% sodium deoxycholate, 2 mM Na3VO4, 25 mM NaF, and a protease inhibitor mixture (1 tablet/10 mL) (Roche)]. After sonication and incubation for 20 min on ice, the extracts were centrifuged at 16,100 × g and 4 °C for 12 min, and the supernatants were collected and stored for protein quantification and Western blotting.
SDS/PAGE and Western Blotting.
The protein concentrations of whole-cell extracts were quantified by the Bradford method (Bio-Rad), measuring the absorbance of the samples at 595 nm on a microplate reader (Multiskan EX; Thermo Scientific). Different concentrations of BSA were used to obtain a standard curve from which the protein concentration of the samples was extrapolated. Unless stated otherwise, SDS sample buffer (Invitrogen) with 70 mM DTT was added to the protein samples (15–30 μg); samples were heated at 95 °C for 5 min and then were loaded onto 10-well 12% polyacrylamide Bis-Tris gels or 15-well 4–12% polyacrylamide Bis-Tris gels (Invitrogen). Samples at RT were separated by electrophoresis at a constant voltage of 100 V over 2.5 h in 3-(N-morpholino)propanesulfonic acid (Mops) SDS running buffer (Invitrogen), and the proteins were then transferred to nitrocellulose membranes (Bio-Rad) for 1 h at 30 V and 4 °C in transfer buffer containing 10% methanol (Panreac), 150 mM glycine (Sigma), and 25 mM Tris (Sigma). The membranes were blocked for 40 min at RT with 5% nonfat milk and/or 5% BSA (Sigma) in Tris-buffered saline with Tween 20 (Sigma) (TBST) buffer [20 mM Tris⋅HCl (pH 7.4), 150 mM NaCl, 0.05% Tween-20], and after washing they were probed overnight at 4 °C with specific primary antibodies (see Antibodies below) dissolved in blocking solution. The membranes were then washed three times for 15 min with TBST and incubated for 2 h at RT with the specific secondary antibodies dissolved in blocking solution (see Antibodies below). Finally, membranes were washed twice for 15 min with TBST and once for 10 min with TBS alone. The bands specific to the proteins of interest were visualized with the Lumi-light plus detection kit using Odyssey Fc (LI-COR), and the proteins were quantified with ImageJ software.
Native PAGE.
Native PAGE samples were prepared in loading buffer [1 M Tris⋅HCl (pH 6.8), glycerol 100% (Panreac), 0.4% bromophenol blue (Sigma)] and loaded without heating or DTT addition in 10% polyacrylamide Tris gels without SDS. Native PAGE samples were separated by electrophoresis with a constant voltage of 80 V during 5 h at 4 °C in two running buffers in independent compartments: cathode buffer (100 mM l-histidine, pH 8.0) and anode buffer (100 mM Tris⋅HCl, pH 8.8). Transference onto nitrocellulose membranes and further steps were performed as for SDS/PAGE Western blotting described above.
Drug Treatments.
A specific inhibitor of PLK2 (49) was synthesized by WuXi AppTec. For survival experiments, a single dose of the PLK2 inhibitor (0.1, 0.5, or 1 μM), or the highest equivalent dose of DMSO (Sigma) as a control, was added to the neurons 1 dpt. To evaluate PLK2 inhibition, neurons were fixed at different times (1–8 dpt), and PS129 was quantified by immunofluorescence. Amlexanox (Sigma) was used as an inhibitor of GRK5 (50) in the primary neuronal cultures. For survival experiments, a single dose of amlexanox (10, 50, or 100 μM), or the highest equivalent dose of DMSO as a control, was added to the neurons 1 dpt. The effect of GRK5 inhibition on PS129 levels was evaluated by immunofluorescence in neurons fixed 48 h later.
Immunofluorescence.
For immunofluorescence experiments, neuronal cultures were fixed 3 dpt unless stated otherwise, by replacing the medium with 4% PFA (Panreac) and 4% sucrose dissolved in PBS. After 8 min the neurons were washed twice with PBS to remove PFA, and they were kept at 4 °C until immunostaining. To ensure antibody entry into the cytoplasm, the neurons were permeabilized with 0.1% Tx-100 in PBS (PBT) for 20 min at RT, and they were then incubated for 20 min in 1 M glycine (BioRad) at RT. For certain antibodies the cells were again fixed for 10 min in ice-cold 100% methanol (Panreac) after PFA and 4% sucrose fixation, followed by two washes in PBS. Nonspecific binding was then blocked for 1 h with 3% goat serum and 3% BSA (Merck-Millipore) dissolved in PBT, and the neurons were incubated for 2 h with the specific primary antibodies dissolved in blocking solution at RT or 4 °C, depending on the antibody used (see Antibodies below). After three washes in PBS, the neurons were the incubated for 2 h at RT with the corresponding secondary antibodies diluted in blocking solution (see Antibodies below), protected from light, and they were finally washed three times with PBS. For DAPI (Sigma) staining the cells were incubated for a further 5 min with DAPI diluted 1:50,000 in PBS, and they were then washed three times in PBS. The coverslips were finally removed from the 24-well plates and placed on microscope slides on an 8-μL drop of mounting medium (25 mg of 1,4-diazabicyclo[2.2.2]octane) (DABCO; Sigma) per milliliter of Permafluor (Thermo Scientific)]. The preparations were dried for 30 min at 37 °C and kept at 4 °C until they were visualized. Images were obtained with a 63× objective on a Zeiss Axiovert 200M fluorescence microscope, and their acquisition and processing were performed using MetaMorph Microscopy Automation and Image Analysis Software (Molecular Devices).
Automated Image Acquisition.
Taking advantage of the nonproliferative nature of neurons, transient transfections with fluorescent-tagged proteins allow us to track neuronal cultures longitudinally over long periods of time (29). Primary cortical neurons growing in 24-well plates were placed on a Zeiss Observer Z1 microscope equipped with a chamber that maintains stable temperature and CO2 at 37 °C and 5% CO2 (Zeiss). Zen System software (Zeiss) allows images to be automatically acquired at determined positions with 10× or 20× long-distance objectives in one or different channels, designating those positions with particular spatial coordinates. A sequential and automated repetition of a series of tasks (locating a particular neuronal field, automatic focusing and image acquisition, and moving forward to the next nonoverlapping neuronal field) permits fast and efficient scanning of multiple neuronal fields per plate. Once the full set of images has been acquired, the plate is returned to the incubator until a new set of images is needed. For a typical survival experiment, 10 positions per well and four wells per condition were used. Positions were chosen randomly, making the selection of neurons to be analyzed unbiased. A template with the same initial spatial positions is used throughout the experiment, following exactly the same neuronal fields. For photoswitching experiments, 100 images per condition were acquired with the 20× objective at manually selected positions. For non–cell-autonomous experiments, a tiled image was created from adjacent nonoverlapping positions covering 24% of the well surface for each condition. For survival experiments, images were acquired with the 10× objective.
Confocal Imaging.
Confocal images were acquired with a LSM-800 confocal microscope (Zeiss) with Zen System 2.3 software (Zeiss) at 63× augmentation. Each confocal image was made of 20–40 Z-stacks, one every 0.31 μm. Orthogonal views and 3D reconstructions were done with Volocity 6.1 software.
Image Processing and Statistics.
Fluorescence intensity was quantified in single neurons with MetaMorph Analysis software. The final average fluorescence intensity values were determined by subtracting the background signal from the fluorescence intensity measured in an area corresponding to the neuronal soma. When measuring fluorescence intensities from multiple channels, the same selected areas were transferred between the images corresponding to each channel. GraphPad Prism 5 software was used to perform Kruskal–Wallis and Dunn’s post hoc tests, one-way ANOVA and Bonferroni’s post hoc tests, and correlation tests, and to obtain the graphs.
For survival experiments, MATLAB-based semiautomated ad hoc programs were developed for image analysis and to estimate the survival times of individual neurons. The program shows the same neuronal field sequentially at all the different experimental times. At the initial time, the neurons were selected and numbered. In the rest of the time series, the software opens each image, and the user compares it with the previous time. Dead neurons are identified and categorized as uncensored events. Neurons that survive until the end of the experiment are considered censored events. The data were exported to Excel, and all further survival analyses were performed with STATA 12.
The Nelson–Aalen cumulative hazard function was used to plot the differences in the cumulative risk of death among the experimental groups. These differences were analyzed with a log-rank test and clustered Cox regression models, as long as a proportional hazard assumption was fulfilled (Schoenfeld residual-based test evaluation or graphical assessment). Otherwise, the variable experimental group was evaluated as a time-varying covariate. Since transfection conditions are not controlled at the level of the individual neuron, a clustered Cox regression analysis of neurons coexisting in the same well was performed to improve the accuracy of the test. In addition, and because all experiments were repeated at least twice, the variability in the baseline toxicity between experiments was adjusted by stratifying the Cox model for each experiment, as long as there was more than one well per condition in any experiment. In experiments with one well per condition in all of the grouped experiments, well and experiment become equivalent, so clustering the wells was sufficient, and no stratification was applied. When necessary, spatial coordinates were assigned to each individual neuron in the culture using custom MATLAB software. Based on this spatial distribution, the number of neurons in an empirically determined radius around each neuron was calculated (nr). Neurons in which the distance to the border of the tiled image was less than the studied radius were excluded from this particular analysis because the data became incomplete. Interaction effects between the number of neurons in a determined radius (nr) and experimental groups were also included in the Cox regression model, as long as the effect of a given variable differed among the distinct experimental groups (Wald test, P < 0.05).
To analyze the photoswitching experiments, specific MATLAB-based custom software was designed inspired by particle-tracking programs. The user selects each neuron at the initial time, and the program automatically detects those neurons at subsequent times, measuring the fluorescence intensity in the green channel. To this end, the image is binarized by applying a threshold, and several morphological operations are performed to determine the pixels that correspond to each neuron. The centroid of the neuron at a given time is then obtained and used to look for the presence of the neuron (in the same region) at the following time point. This is possible because neurons remain at roughly the same positions throughout the experiment. Additionally, the pixels corresponding to each neuron (obtained from the green channel) are used to calculate the average red fluorescence in the neuron (red channel). From these data the program adjusts the average RFI for every single neuron to an exponential decay over time. In practical terms, we perform a logarithmic transformation of the intensity values and apply a linear regression, obtaining the slope (−K) and the coefficient of determination (R2). The t1/2 is therefore calculated for each neuron with the following equation t1/2 = −LN (2)/K.
Antibodies.
Total aSyn: BD Transduction Laboratories (610786). Western blot (WB) (1:1,000), immunofluorescence (IF) (1:100)
Total aSyn: Abcam (ab24717). WB (1:1,000), IF (1:200)
Total aSyn: Synaptic Systems (128211). IF (1:100)
PS129 aSyn: Abcam (ab168381). WB (1:1,000), IF (1:1,000)
Tubulin: Sigma (T6199). WB (1:10,000)
Vimentin: Sigma (V6389). WB (1:5,000)
ECL anti-mouse IgG HRP linked: GE Healthcare (NA931). WB (1:15,000)
ECL anti-rabbit IgG HRP linked: GE Healthcare (NA934). WB (1:15,000)
FITC-conjugated anti-mouse IgG: Jackson ImmunoResearch (115-095-003). IF (1:200)
Cy5-conjugated anti-mouse IgG: Jackson ImmunoResearch (115-175-166). IF (1:500)
Cy3-conjugated anti-mouse IgG: Jackson ImmunoResearch (115-165-003). IF (1:500)
Cy5-conjugated anti-rabbit IgG: Jackson ImmunoResearch (115-175-144). IF (1:500)
mCherry: Abcam (Ab125096). WB (1:2,000), IF (1:500)
GFP: Life Technologies (A11122). WB (1:1,000), IF (1:200)
Dendra2: OriGene (TA180094). WB (1:500), IF (1:100)
GRK5: Millipore (05-466). WB (1:1,000)
APP: BioLegend 6E10 (803001). IF (1:1,000)
Acknowledgments
We thank R. Edwards for the empty pCAGGs vector and GFP–α-synuclein fusions (WT/A30P/A53T); J. Wesseling for the pSUPER-mCherry; T. Südhof for the lentiviral expression plasmids (GFP, myc-Syn, and myc-E46KSyn); S. Knafo for the APPswe/lnd expression plasmid; R. Hernández for the pIRES plasmid; I. Pérez-Otaño, J. Wesseling, and T. Aragón for critical reading of the manuscript; and T. Aragón for scientific discussions and support. This work was supported by the European Commission’s Seventh Framework Program FP7/Marie Curie Reintegration Grant 224849 (to M.A.), Ministerio de Economía y Competitividad, Spain Grants RYC2008-03254 (to M.A.), BFU2012-39737 (to M.A.), BFU2013-48703P (to M.A.), and IEDI2015-00610 (to M.A.); the Fundación Tatiana P. de Guzmán El Bueno (M.A.), and the Fundación para la Investigación Médica Aplicada (FIMA). I.Í.-M. and R.B. were supported by FIMA and Asociación de Amigos (ADA) Predoctoral Fellowships from the University of Navarra, respectively.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. T.F.O. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703420114/-/DCSupplemental.
References
- 1.Halliday GM, Holton JL, Revesz T, Dickson DW. Neuropathology underlying clinical variability in patients with synucleinopathies. Acta Neuropathol. 2011;122:187–204. doi: 10.1007/s00401-011-0852-9. [DOI] [PubMed] [Google Scholar]
- 2.Singleton AB, Farrer MJ, Bonifati V. The genetics of Parkinson’s disease: Progress and therapeutic implications. Mov Disord. 2013;28:14–23. doi: 10.1002/mds.25249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Satake W, et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson’s disease. Nat Genet. 2009;41:1303–1307. doi: 10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- 4.Nalls MA, et al. International Parkinson’s Disease Genomics Consortium (IPDGC); Parkinson’s Study Group (PSG) Parkinson’s Research: The Organized GENetics Initiative (PROGENI); 23andMe; GenePD; NeuroGenetics Research Consortium (NGRC); Hussman Institute of Human Genomics (HIHG); Ashkenazi Jewish Dataset Investigator; Cohorts for Health and Aging Research in Genetic Epidemiology (CHARGE); North American Brain Expression Consortium (NABEC); United Kingdom Brain Expression Consortium (UKBEC); Greek Parkinson’s Disease Consortium; Alzheimer Genetic Analysis Group Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat Genet. 2014;46:989–993. doi: 10.1038/ng.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maraganore DM, et al. Genetic Epidemiology of Parkinson’s Disease (GEO-PD) Consortium Collaborative analysis of alpha-synuclein gene promoter variability and Parkinson disease. JAMA. 2006;296:661–670. doi: 10.1001/jama.296.6.661. [DOI] [PubMed] [Google Scholar]
- 6.Soldner F, et al. Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature. 2016;533:95–99. doi: 10.1038/nature17939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Theillet FX, et al. Structural disorder of monomeric α-synuclein persists in mammalian cells. Nature. 2016;530:45–50. doi: 10.1038/nature16531. [DOI] [PubMed] [Google Scholar]
- 8.Bartels T, Choi JG, Selkoe DJ. α-Synuclein occurs physiologically as a helically folded tetramer that resists aggregation. Nature. 2011;477:107–110. doi: 10.1038/nature10324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cremades N, et al. Direct observation of the interconversion of normal and toxic forms of α-synuclein. Cell. 2012;149:1048–1059. doi: 10.1016/j.cell.2012.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts HL, Brown DR. Seeking a mechanism for the toxicity of oligomeric α-synuclein. Biomolecules. 2015;5:282–305. doi: 10.3390/biom5020282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spillantini MG, et al. Alpha-synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 12.Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- 13.Ono K, Ikeda T, Takasaki J, Yamada M. Familial Parkinson disease mutations influence α-synuclein assembly. Neurobiol Dis. 2011;43:715–724. doi: 10.1016/j.nbd.2011.05.025. [DOI] [PubMed] [Google Scholar]
- 14.Fujiwara H, et al. Alpha-synuclein is phosphorylated in synucleinopathy lesions. Nat Cell Biol. 2002;4:160–164. doi: 10.1038/ncb748. [DOI] [PubMed] [Google Scholar]
- 15.Oueslati A. Implication of alpha-synuclein phosphorylation at S129 in synucleinopathies: What have we learned in the last decade? J Parkinsons Dis. 2016;6:39–51. doi: 10.3233/JPD-160779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brettschneider J, Del Tredici K, Lee VM, Trojanowski JQ. Spreading of pathology in neurodegenerative diseases: A focus on human studies. Nat Rev Neurosci. 2015;16:109–120. doi: 10.1038/nrn3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mollenhauer B, et al. α-synuclein in human cerebrospinal fluid is principally derived from neurons of the central nervous system. J Neural Transm (Vienna) 2012;119:739–746. doi: 10.1007/s00702-012-0784-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tokuda T, et al. Detection of elevated levels of α-synuclein oligomers in CSF from patients with Parkinson disease. Neurology. 2010;75:1766–1772. doi: 10.1212/WNL.0b013e3181fd613b. [DOI] [PubMed] [Google Scholar]
- 19.Emmanouilidou E, et al. Cell-produced alpha-synuclein is secreted in a calcium-dependent manner by exosomes and impacts neuronal survival. J Neurosci. 2010;30:6838–6851. doi: 10.1523/JNEUROSCI.5699-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Danzer KM, et al. Exosomal cell-to-cell transmission of alpha synuclein oligomers. Mol Neurodegener. 2012;7:42. doi: 10.1186/1750-1326-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Desplats P, et al. Inclusion formation and neuronal cell death through neuron-to-neuron transmission of alpha-synuclein. Proc Natl Acad Sci USA. 2009;106:13010–13015. doi: 10.1073/pnas.0903691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Volpicelli-Daley LA, et al. Exogenous α-synuclein fibrils induce Lewy body pathology leading to synaptic dysfunction and neuron death. Neuron. 2011;72:57–71. doi: 10.1016/j.neuron.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luk KC, et al. Pathological α-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science. 2012;338:949–953. doi: 10.1126/science.1227157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osterberg VR, et al. Progressive aggregation of alpha-synuclein and selective degeneration of lewy inclusion-bearing neurons in a mouse model of parkinsonism. Cell Rep. 2015;10:1252–1260. doi: 10.1016/j.celrep.2015.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helwig M, et al. Brain propagation of transduced α-synuclein involves non-fibrillar protein species and is enhanced in α-synuclein null mice. Brain. 2016;139:856–870. doi: 10.1093/brain/awv376. [DOI] [PubMed] [Google Scholar]
- 26.Braak H, et al. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol Aging. 2003;24:197–211. doi: 10.1016/s0197-4580(02)00065-9. [DOI] [PubMed] [Google Scholar]
- 27.Walsh DM, Selkoe DJ. A critical appraisal of the pathogenic protein spread hypothesis of neurodegeneration. Nat Rev Neurosci. 2016;17:251–260. doi: 10.1038/nrn.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Surmeier DJ, Obeso JA, Halliday GM. Selective neuronal vulnerability in Parkinson disease. Nat Rev Neurosci. 2017;18:101–113. doi: 10.1038/nrn.2016.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arrasate M, Finkbeiner S. Automated microscope system for determining factors that predict neuronal fate. Proc Natl Acad Sci USA. 2005;102:3840–3845. doi: 10.1073/pnas.0409777102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 31.Miller J, et al. Identifying polyglutamine protein species in situ that best predict neurodegeneration. Nat Chem Biol. 2011;7:925–934. doi: 10.1038/nchembio.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skibinski G, Nakamura K, Cookson MR, Finkbeiner S. Mutant LRRK2 toxicity in neurons depends on LRRK2 levels and synuclein but not kinase activity or inclusion bodies. J Neurosci. 2014;34:418–433. doi: 10.1523/JNEUROSCI.2712-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barmada SJ, et al. Amelioration of toxicity in neuronal models of amyotrophic lateral sclerosis by hUPF1. Proc Natl Acad Sci USA. 2015;112:7821–7826. doi: 10.1073/pnas.1509744112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsvetkov AS, et al. Proteostasis of polyglutamine varies among neurons and predicts neurodegeneration. Nat Chem Biol. 2013;9:586–592. doi: 10.1038/nchembio.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skibinski G, et al. Nrf2 mitigates LRRK2- and α-synuclein-induced neurodegeneration by modulating proteostasis. Proc Natl Acad Sci USA. 2017;114:1165–1170. doi: 10.1073/pnas.1522872114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohrfelt A, et al. Identification of novel α-synuclein isoforms in human brain tissue by using an online nanoLC-ESI-FTICR-MS method. Neurochem Res. 2011;36:2029–2042. doi: 10.1007/s11064-011-0527-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez-Vicente M, et al. Dopamine-modified alpha-synuclein blocks chaperone-mediated autophagy. J Clin Invest. 2008;118:777–788. doi: 10.1172/JCI32806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gurskaya NG, et al. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat Biotechnol. 2006;24:461–465. doi: 10.1038/nbt1191. [DOI] [PubMed] [Google Scholar]
- 39.Okochi M, et al. Constitutive phosphorylation of the Parkinson’s disease associated alpha-synuclein. J Biol Chem. 2000;275:390–397. doi: 10.1074/jbc.275.1.390. [DOI] [PubMed] [Google Scholar]
- 40.Miake H, Mizusawa H, Iwatsubo T, Hasegawa M. Biochemical characterization of the core structure of alpha-synuclein filaments. J Biol Chem. 2002;277:19213–19219. doi: 10.1074/jbc.M110551200. [DOI] [PubMed] [Google Scholar]
- 41.Burré J, Sharma M, Südhof TC. Systematic mutagenesis of α-synuclein reveals distinct sequence requirements for physiological and pathological activities. J Neurosci. 2012;32:15227–15242. doi: 10.1523/JNEUROSCI.3545-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsika E, et al. Distinct region-specific alpha-synuclein oligomers in A53T transgenic mice: Implications for neurodegeneration. J Neurosci. 2010;30:3409–3418. doi: 10.1523/JNEUROSCI.4977-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mbefo MK, et al. Parkinson disease mutant E46K enhances α-synuclein phosphorylation in mammalian cell lines, in yeast, and in vivo. J Biol Chem. 2015;290:9412–9427. doi: 10.1074/jbc.M114.610774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bergeron M, et al. In vivo modulation of polo-like kinases supports a key role for PLK2 in Ser129 α-synuclein phosphorylation in mouse brain. Neuroscience. 2014;256:72–82. doi: 10.1016/j.neuroscience.2013.09.061. [DOI] [PubMed] [Google Scholar]
- 45.Arawaka S, et al. The role of G-protein-coupled receptor kinase 5 in pathogenesis of sporadic Parkinson’s disease. J Neurosci. 2006;26:9227–9238. doi: 10.1523/JNEUROSCI.0341-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seeburg DP, Feliu-Mojer M, Gaiottino J, Pak DT, Sheng M. Critical role of CDK5 and polo-like kinase 2 in homeostatic synaptic plasticity during elevated activity. Neuron. 2008;58:571–583. doi: 10.1016/j.neuron.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matsumoto T, et al. Polo-like kinases mediate cell survival in mitochondrial dysfunction. Proc Natl Acad Sci USA. 2009;106:14542–14546. doi: 10.1073/pnas.0904229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Michal AM, et al. G Protein-coupled receptor kinase 5 is localized to centrosomes and regulates cell cycle progression. J Biol Chem. 2012;287:6928–6940. doi: 10.1074/jbc.M111.298034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bowers S, et al. Design and synthesis of highly selective, orally active polo-like kinase-2 (Plk-2) inhibitors. Bioorg Med Chem Lett. 2013;23:2743–2749. doi: 10.1016/j.bmcl.2013.02.065. [DOI] [PubMed] [Google Scholar]
- 50.Homan KT, Wu E, Cannavo A, Koch WJ, Tesmer JJ. Identification and characterization of amlexanox as a G protein-coupled receptor kinase 5 inhibitor. Molecules. 2014;19:16937–16949. doi: 10.3390/molecules191016937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nemani VM, et al. Increased expression of alpha-synuclein reduces neurotransmitter release by inhibiting synaptic vesicle reclustering after endocytosis. Neuron. 2010;65:66–79. doi: 10.1016/j.neuron.2009.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Selkoe DJ, Hardy J. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol Med. 2016;8:595–608. doi: 10.15252/emmm.201606210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Knafo S, et al. PTEN recruitment controls synaptic and cognitive function in Alzheimer’s models. Nat Neurosci. 2016;19:443–453. doi: 10.1038/nn.4225. [DOI] [PubMed] [Google Scholar]
- 54.Kasten M, Klein C. The many faces of alpha-synuclein mutations. Mov Disord. 2013;28:697–701. doi: 10.1002/mds.25499. [DOI] [PubMed] [Google Scholar]
- 55.Zarranz JJ, et al. The new mutation, E46K, of alpha-synuclein causes Parkinson and Lewy body dementia. Ann Neurol. 2004;55:164–173. doi: 10.1002/ana.10795. [DOI] [PubMed] [Google Scholar]
- 56.Conway KA, Harper JD, Lansbury PT. Accelerated in vitro fibril formation by a mutant alpha-synuclein linked to early-onset Parkinson disease. Nat Med. 1998;4:1318–1320. doi: 10.1038/3311. [DOI] [PubMed] [Google Scholar]
- 57.Outeiro TF, Lindquist S. Yeast cells provide insight into alpha-synuclein biology and pathobiology. Science. 2003;302:1772–1775. doi: 10.1126/science.1090439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fortin DL, et al. Lipid rafts mediate the synaptic localization of alpha-synuclein. J Neurosci. 2004;24:6715–6723. doi: 10.1523/JNEUROSCI.1594-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Emmer KL, Waxman EA, Covy JP, Giasson BI. E46K human alpha-synuclein transgenic mice develop Lewy-like and tau pathology associated with age-dependent, detrimental motor impairment. J Biol Chem. 2011;286:35104–35118. doi: 10.1074/jbc.M111.247965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cannon JR, et al. Expression of human E46K-mutated α-synuclein in BAC-transgenic rats replicates early-stage Parkinson’s disease features and enhances vulnerability to mitochondrial impairment. Exp Neurol. 2013;240:44–56. doi: 10.1016/j.expneurol.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee Y, Dawson VL, Dawson TM. Animal models of Parkinson’s disease: Vertebrate genetics. Cold Spring Harb Perspect Med. 2012;2:a009324. doi: 10.1101/cshperspect.a009324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Richfield EK, et al. Behavioral and neurochemical effects of wild-type and mutated human alpha-synuclein in transgenic mice. Exp Neurol. 2002;175:35–48. doi: 10.1006/exnr.2002.7882. [DOI] [PubMed] [Google Scholar]
- 63.Nuber S, et al. Neurodegeneration and motor dysfunction in a conditional model of Parkinson’s disease. J Neurosci. 2008;28:2471–2484. doi: 10.1523/JNEUROSCI.3040-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Janezic S, et al. Deficits in dopaminergic transmission precede neuron loss and dysfunction in a new Parkinson model. Proc Natl Acad Sci USA. 2013;110:E4016–E4025. doi: 10.1073/pnas.1309143110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kramer ML, Schulz-Schaeffer WJ. Presynaptic alpha-synuclein aggregates, not Lewy bodies, cause neurodegeneration in dementia with Lewy bodies. J Neurosci. 2007;27:1405–1410. doi: 10.1523/JNEUROSCI.4564-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Milber JM, et al. Lewy pathology is not the first sign of degeneration in vulnerable neurons in Parkinson disease. Neurology. 2012;79:2307–2314. doi: 10.1212/WNL.0b013e318278fe32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Berg D, et al. Time to redefine PD? Introductory statement of the MDS task force on the definition of Parkinson’s disease. Mov Disord. 2014;29:454–462. doi: 10.1002/mds.25844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Villar-Piqué A, et al. Environmental and genetic factors support the dissociation between α-synuclein aggregation and toxicity. Proc Natl Acad Sci USA. 2016;113:E6506–E6515. doi: 10.1073/pnas.1606791113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Winner B, et al. In vivo demonstration that alpha-synuclein oligomers are toxic. Proc Natl Acad Sci USA. 2011;108:4194–4199. doi: 10.1073/pnas.1100976108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McCormack AL, Mak SK, Di Monte DA. Increased α-synuclein phosphorylation and nitration in the aging primate substantia nigra. Cell Death Dis. 2012;3:e315. doi: 10.1038/cddis.2012.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Samuel F, et al. Effects of serine 129 phosphorylation on α-synuclein aggregation, membrane association, and internalization. J Biol Chem. 2016;291:4374–4385. doi: 10.1074/jbc.M115.705095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oueslati A, Schneider BL, Aebischer P, Lashuel HA. Polo-like kinase 2 regulates selective autophagic α-synuclein clearance and suppresses its toxicity in vivo. Proc Natl Acad Sci USA. 2013;110:E3945–E3954. doi: 10.1073/pnas.1309991110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee KJ, et al. Requirement for Plk2 in orchestrated ras and rap signaling, homeostatic structural plasticity, and memory. Neuron. 2011;69:957–973. doi: 10.1016/j.neuron.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wong YC, Krainc D. α-synuclein toxicity in neurodegeneration: Mechanism and therapeutic strategies. Nat Med. 2017;23:1–13. doi: 10.1038/nm.4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tanik SA, Schultheiss CE, Volpicelli-Daley LA, Brunden KR, Lee VM. Lewy body-like α-synuclein aggregates resist degradation and impair macroautophagy. J Biol Chem. 2013;288:15194–15210. doi: 10.1074/jbc.M113.457408. [DOI] [PMC free article] [PubMed] [Google Scholar]