Significance
Extremely acidic environments are scattered worldwide, and their ecosystems are supported by acidophilic microalgae as primary producers. To understand how acidophilic algae evolved from their respective neutrophilic ancestors, we determined the draft genome sequence of the acidophilic green alga Chlamydomonas eustigma and performed comparative genome analyses between C. eustigma and its neutrophilic relative Chlamydomonas reinhardtii. The results suggest that higher expression of heat-shock proteins and H+-ATPase, loss of some metabolic pathways that acidify cytosol, and acquisition of metal-detoxifying genes by horizontal gene transfer have played important roles in the adaptation to acidic environments. These features are also found in other acidophilic green and red algae, suggesting the existence of common mechanisms in the adaptation to acidic environments.
Keywords: environmental adaptation, acidic environment, acidophilic alga, comparative genomics, comparative transcriptomics
Abstract
Some microalgae are adapted to extremely acidic environments in which toxic metals are present at high levels. However, little is known about how acidophilic algae evolved from their respective neutrophilic ancestors by adapting to particular acidic environments. To gain insights into this issue, we determined the draft genome sequence of the acidophilic green alga Chlamydomonas eustigma and performed comparative genome and transcriptome analyses between C. eustigma and its neutrophilic relative Chlamydomonas reinhardtii. The results revealed the following features in C. eustigma that probably contributed to the adaptation to an acidic environment. Genes encoding heat-shock proteins and plasma membrane H+-ATPase are highly expressed in C. eustigma. This species has also lost fermentation pathways that acidify the cytosol and has acquired an energy shuttle and buffering system and arsenic detoxification genes through horizontal gene transfer. Moreover, the arsenic detoxification genes have been multiplied in the genome. These features have also been found in other acidophilic green and red algae, suggesting the existence of common mechanisms in the adaptation to acidic environments.
Several eukaryotic microalgae have been identified in acidic environments (pH <4.0) such as acid mine drainage (AMD) and geothermal hot springs (1). In this pH range, cyanobacteria are not present, and only acidophilic eukaryotic phototrophs are capable of photosynthesis (Fig. 1) (2, 3). The extremely low pH of these waters is due to the dissolution and oxidation of sulfur that is exposed to water and oxygen and produces sulfuric acid (4). The low pH facilitates metal solubility in water; therefore, acidic waters tend to have high concentrations of metals (5). Thus, acidophilic eukaryotic algae usually possess the ability to cope with toxic heavy metals in addition to low pH, both of which are lethal to most eukaryotes (2). Acidophilic algae are distributed throughout different branches of the eukaryotes, such as in red and green algae, stramenopiles, and euglenids. In most cases, neutrophilic relatives have been identified, suggesting that acidophilic algae evolved from their respective neutrophilic ancestors multiple times independently (6). However, it is largely unknown how several lineages of algae have successfully adapted to their acidic environments.
Fig. 1.
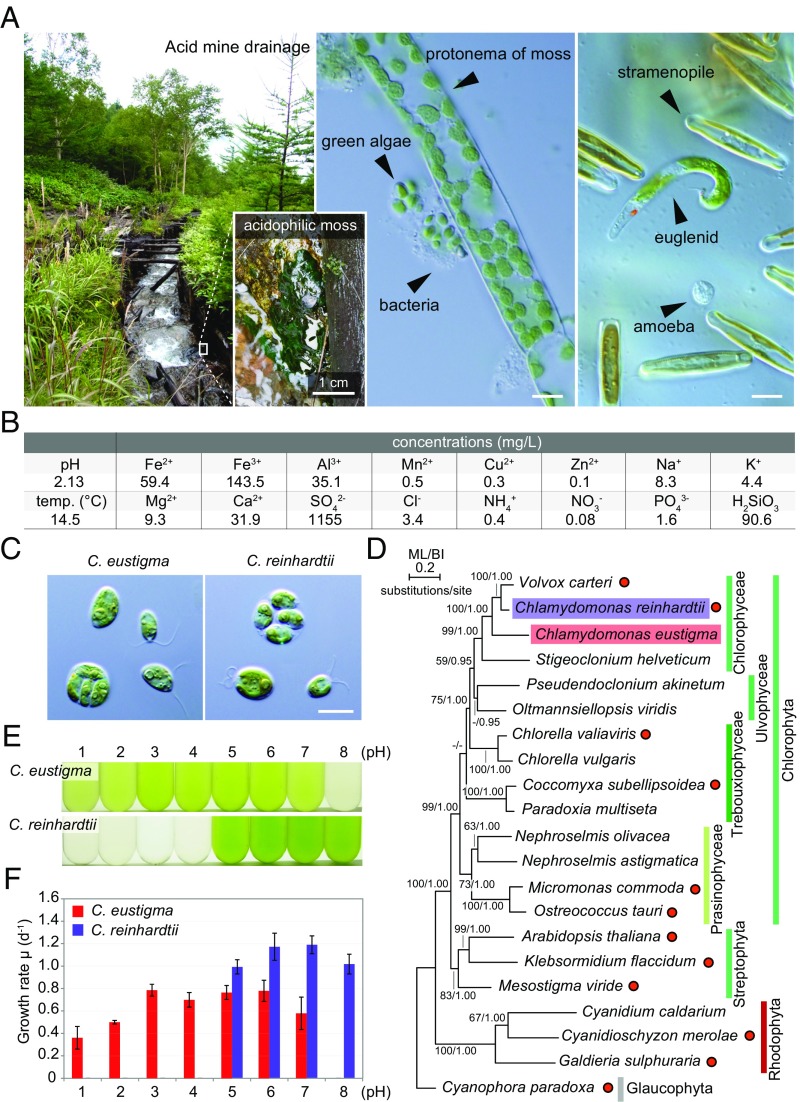
Habitat, taxonomic position, and physiological features of the acidophilic green alga C. eustigma. (A) The algae inhabiting AMD in Yokote, Nagano Prefecture, Japan, and confirmation of the existence of C. eustigma. Algae were found predominantly in association with acidophilic mosses. (Scale bars: 10 μm.) (B) pH, temperature, and concentrations of some ions in the AMD. (C) Cells of C. eustigma NIES-2499 (Left) and C. reinhardtii 137c mt+ (Right). (Scale bar: 10 µm.) (D) A phylogenetic tree of green and red algae based on the concatenated datasets (21 taxa, 11,367 sites) of five chloroplast protein-coding genes (atpB, psaA, psaB, psbC, and rbcL) and chloroplast ribosomal DNA sequence (16S and 23S). The maximum likelihood (ML) (RaxML 8.0.0) and Bayesian (MrBayes 3.2.6) analyses were calculated under separate model conditions. Bootstrap values (BP) >50% obtained by ML and Bayesian posterior probabilities (BI) >0.95 obtained by Bayesian analysis (MrBayes 3.2.6) are shown above the branches. The branch lengths reflect the evolutionary distances indicated by the scale bar. Filled red circles on the right indicate organisms for which genomes have been sequenced thus far. (E) C. eustigma and C. reinhardtii were cultured for 1 d in the same photoautotrophic medium at a series of pH. (F) Growth rates of C. eustigma and C. reinhardtii based on the increase in the cell number at the indicated pH. The error bars represent the SD of three biological replicates.
Thus, far, the genomes of three related thermo-acidophilic red algae, Cyanidioschyzon merolae (7), Galdieria sulphuraria (8), and Galdieria phlegrea (9), have been sequenced (all belong to the cyanidialean red algae, which inhabit sulfuric hot springs worldwide and grow optimally at 40–45 °C and pH 2–3). Genomic analyses showed that horizontal gene transfer (HGT) from environmental prokaryotes, the expansion of gene families, and the loss of genes have probably played important roles in the adaptation of Cyanidiales to acidic and high-temperature environments (8). Through HGT, cyanidialean red algae acquired arsenical-resistance efflux pumps that biotransform arsenic and archaeal ATPases, which probably contribute to the algal heat tolerance (8). In addition, the reduction in the number of genes encoding voltage-gated ion channels and the expansion of chloride channel and chloride carrier/channel families in the genome has probably contributed to the algal acid tolerance (8). Likewise, a study in the acidophilic green alga Chlamydomonas acidophila showed that phytochelatin synthase genes of bacterial HGT origin played an important role in the tolerance to cadmium (10).
However, the genomes of acidophilic algae other than cyanidialean red algae have not been sequenced. The green and red algae diverged relatively soon after the emergence of primitive eukaryotic algae (11). In addition, comparisons with neutrophilic relatives are feasible in the case of acidophilic green algae but are difficult in the case of cyanidialean red algae because their last common acidophilic ancestor diverged from other neutrophilic red algae 1.2–1.3 billion y ago (12). Thus, whole-genome comparisons between evolutionarily related neutrophilic and acidophilic green algae will give insights into how acidophiles evolved from their neutrophilic ancestors.
Here, we determined the draft genome of the acidophilic green alga Chlamydomonas eustigma NIES-2499 isolated from sulfuric AMD and performed comparative genome and transcriptome analyses between C. eustigma and its neutrophilic relative Chlamydomonas reinhardtii, which was previously fully sequenced (13). The results suggest that up-regulation of genes encoding heat-shock proteins (HSPs) and plasma membrane H+-ATPase (PMA), loss of fermentative genes that produce organic acids and thus reduce cytosolic pH, the acquisition of an energy shuttle and buffering system, and the acquisition and multiplication of genes involved in arsenic biotransformation and detoxification have contributed to the adaptation of C. eustigma to acidic conditions. The results also suggest that there are several commonalities in genomic evolution for adapting to acidic environments among red algae and green algae.
Results
Habitat, Taxonomic Position, and Physiological Features of the Acidophilic Green Alga C. eustigma.
C. eustigma (a haploid vegetative cell) was originally isolated together with mosses (14) from sulfuric AMD in Nagano Prefecture, Japan, in August 1992. We confirmed that C. eustigma still thrived in that AMD (pH 2.13, 14.5 °C) in September 2013 (Fig. 1A). The water in the AMD contained high concentrations of iron, Al3+, and SO42− (Fig. 1B), as in the case of other AMDs (15). C. eustigma exhibits a cell size and morphology very similar to those of the neutrophilic C. reinhardtii. Both cells possess two flagella and a large cup-shaped chloroplast in which an eyespot and a large pyrenoid are formed and proliferate by forming autospores in the mother cell (Fig. 1C). As also shown by a previous phylogenetic study based on 18S rDNA sequences (16), phylogenetic analysis based on five chloroplast-encoded genes and two chloroplast ribosomal DNA sequences (Table S1) showed that C. eustigma, together with the fully sequenced neutrophile C. reinhardtii (13), belongs to the Chlorophyceae, which contains mainly neutrophilic algae (Fig. 1D). In the phylogenetic tree, all the Chlorophyta (green algae) except for C. eustigma are neutrophilic (Fig. 1D), suggesting that the acidophile C. eustigma evolved from a neutrophilic ancestor. In autotrophic synthetic medium at 20 °C, C. eustigma proliferated at pH 2.0–6.0 (at pH 1.0 cells grew for a few days, but after that they died), and pH 3.0–6.0 was optimal for its growth, whereas C. reinhardtii proliferated at pH 5.0–8.0, and pH 6.0–7.0 was optimal for its growth (Fig. 1 E and F).
Table S1.
Taxon sampling for the phylogeny based on five chloroplast-encoded genes and two chloroplast rDNA sequences
| Species | Class | Division | atpB | psaA | psaB | psbC | rbcL | 16S | 23S |
| Volvox carteri | Chlorophyceae | Chlorophyta | GU084820 | GU084820 | GU084820 | GU084820 | GU084820 | GU084820 | GU084820 |
| Chlamydomonas reinhardtii | Chlorophyceae | Chlorophyta | BK000554 | BK000554 | BK000554 | BK000554 | BK000554 | BK000554 | BK000554 |
| Chlamydomonas eustigma | Chlorophyceae | Chlorophyta | LC229070 | LC229069 | LC229071 | LC229072 | LC229073 | LC229075 | LC229074 |
| Stigeoclonium helveticum | Chlorophyceae | Chlorophyta | DQ630521 | DQ630521 | DQ630521 | DQ630521 | DQ630521 | DQ630521 | DQ630521 |
| Pseudendoclonium akinetum | Ulvophyceae | Chlorophyta | AY835431 | AY835431 | AY835431 | AY835431 | AY835431 | AY835431 | AY835431 |
| Oltmannsiellopsis viridis | Ulvophyceae | Chlorophyta | DQ291132 | DQ291132 | DQ291132 | DQ291132 | DQ291132 | DQ291132 | DQ291132 |
| Chlorella variabilis NC64A | Trebouxiophyceae | Chlorophyta | KJ718922 | KJ718922 | KJ718922 | KJ718922 | KJ718922 | KJ718922 | KJ718922 |
| Chlorella vulgaris | Trebouxiophyceae | Chlorophyta | AB001684 | AB001684 | AB001684 | AB001684 | AB001684 | AB001684 | AB001684 |
| Coccomyxa subellipsoidea C-169 | Trebouxiophyceae | Chlorophyta | HQ693844 | HQ693844 | HQ693844 | HQ693844 | HQ693844 | HQ693844 | HQ693844 |
| Paradoxia multiseta | Trebouxiophyceae | Chlorophyta | KM462879 | KM462879 | KM462879 | KM462879 | KM462879 | KM462879 | KM462879 |
| Nephroselmis olivacea | Prasinophyceae | Chlorophyta | AF137379 | AF137379 | AF137379 | AF137379 | AF137379 | AF137379 | AF137379 |
| Nephroselmis astigmatica | Prasinophyceae | Chlorophyta | KJ746600 | KJ746600 | KJ746600 | KJ746600 | KJ746600 | KJ746600 | KJ746600 |
| Micromonas commoda RCC299 | Prasinophyceae | Chlorophyta | FJ858267 | FJ858267 | FJ858267 | FJ858267 | FJ858267 | FJ858267 | FJ858267 |
| Ostreococcus tauri | Prasinophyceae | Chlorophyta | CR954199 | CR954199 | CR954199 | CR954199 | CR954199 | CR954199 | CR954199 |
| Arabidopsis thaliana | Magnoliopsida | Streptophyta | AP000423 | AP000423 | AP000423 | AP000423 | AP000423 | AP000423 | AP000423 |
| Klebsormidium flaccidum | Charophyceae | Streptophyta | DF238762 | DF238762 | DF238762 | DF238762 | DF238762 | DF238762 | DF238762 |
| Mesostigma viride | Mesostigmatophyceae | Streptophyta | AF166114 | AF166114 | AF166114 | AF166114 | AF166114 | AF166114 | AF166114 |
| Cyanidium caldarium RK1 | Cyanidiophyceae | Rhodophyta | AF022186 | AF022186 | AF022186 | AF022186 | AF022186 | AF022186 | AF022186 |
| Cyanidioschyzon merolae 10D | Cyanidiophyceae | Rhodophyta | AB002583 | AB002583 | AB002583 | AB002583 | AB002583 | AB002583 | AB002583 |
| Galdieria sulphuraria 074W | Cyanidiophyceae | Rhodophyta | KJ700459 | KJ700459 | KJ700459 | KJ700459 | KJ700459 | KJ700459 | KJ700459 |
| Cyanophora paradoxa | Glaucophyceae | Glaucophyta | U30821 | U30821 | U30821 | U30821 | U30821 | U30821 | U30821 |
atpB, beta subunit of ATP synthase; psaA, P700 chlorophyll a-apoprotein A1; psaB, P700 chlorophyll a-apoprotein A2; psbC, photosystem II CP43 apoprotein; rbcL, large subunit of rubisco; 16S, 16S ribosomal RNA; 23S, 23S ribosomal RNA.
Characteristics of the C. eustigma Nuclear Genome.
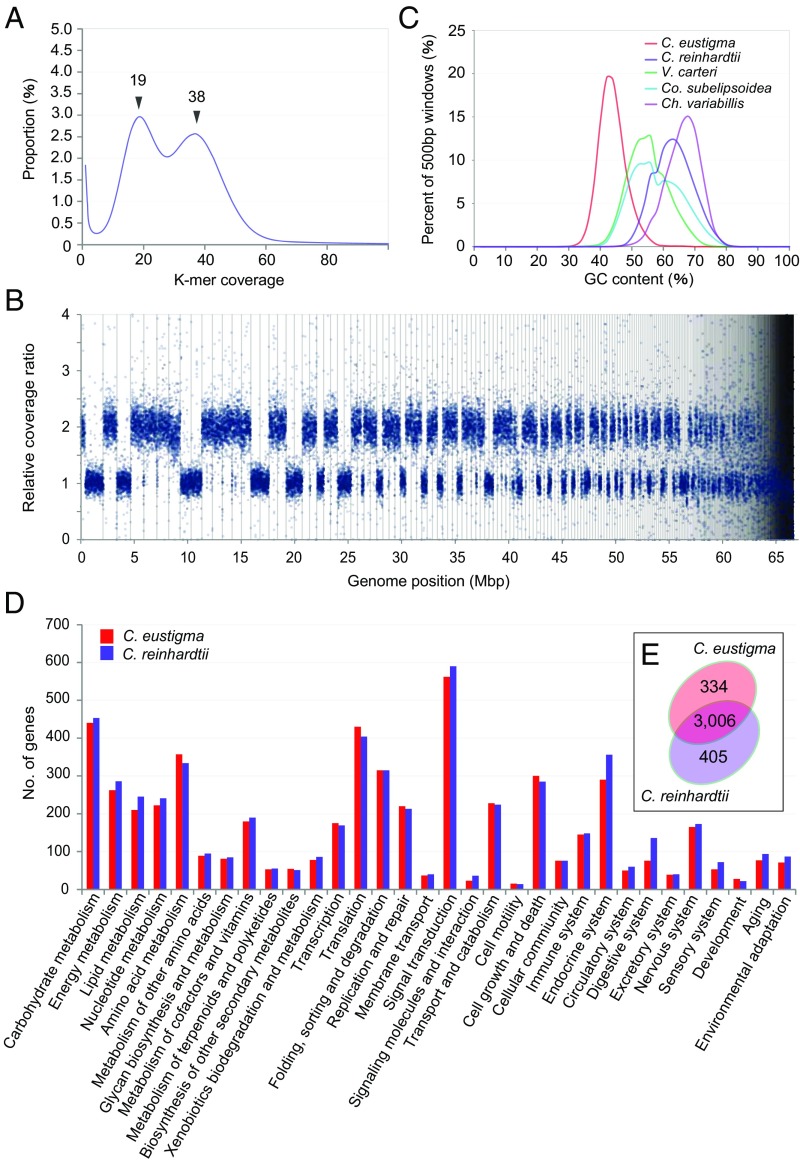
To understand the genetic basis of the adaptation of C. eustigma to an acidic environment, we sequenced its nuclear genome (Tables S2 and S3). K-mer analysis of Illumina MiSeq reads yielded two peaks with similar frequency of coverage (19× and 38×, respectively), suggesting that the C. eustigma genome is a chimera of single and duplicated regions (Fig. 2A). Considering the length of the duplicated regions, the estimated genome size was ∼130 Mb (Table S3). Then we obtained Illumina HiSeq and Roche 454 GS FLX+ reads of the nuclear genome (Table S2). The sequenced DNA reads were assembled into 519 scaffolds (the N50 scaffold size was 465 kb, and the total length was 67 Mb) (Table S3). Consistent with the result of K-mer analysis, sequencing coverage ratios of some scaffolds were two times those of other scaffolds (Fig. 2B and Dataset S1).
Table S2.
Summary of the Roche 454 and Illumina HiSeq and MiSeq data used for this study
| Platform | Libraries | Total sequences, GB | No. of reads | Avg. read length, bp |
| 454 GS FLX+ | Shotgun | 1.45 | 2,436,211 | 596 |
| 8-kb paired end | 0.53 | 1,374,104 | 387 | |
| HiSeq | 400-bp paired end | 2 | 20,000,000 | 100 |
| MiSeq | 800-bp paired end | 2.9 | 13,321,492 | 220 |
| 3-kb mate pair | 0.72 | 5,115,846 | 141 | |
| 5-kb mate pair | 0.59 | 4,068,052 | 145 | |
| 8-kb mate pair | 0.51 | 3,489,420 | 147 |
Table S3.
Summary of statistics for genome-level analyses of C. eustigma and C. reinhardtii
| Features | C. eustigma | C. reinhardtii |
| Genome statistics | ||
| Estimated genome size | ∼130 Mb | — |
| Total bases in scaffolds | 67 Mb (∼110 MB) | 111.1 Mb |
| Genomic G+C content, % | 44.82 | 64.08 |
| No. of assembled scaffolds | 519 | 54 |
| Scaffold N50 size | 465 kb | 7.8 Mb |
| No. of contigs | 3,211 | 1,495 |
| Contig N50 size | 46.2 kb | 219.4 kb |
| Gene statistics | ||
| Predicted number of nuclear genes | 14,105 | 17,741 |
| KEGG annotated genes | 4,470 | 4,741 |
| Average CDS length | 1,745 | 2,207 |
| CDS G+C content, % | 50.57 | 70.24 |
CDS, coding sequence.
Fig. 2.
The C. eustigma genome architecture and comparison of genome contents between C. eustigma and C. reinhardtii. (A) 31 K-mer depth distribution of whole-genome Illumina MiSeq reads. Two peaks at 19× and 38× were identified. (B) Distribution of the relative sequencing coverage ratio in the C. eustigma genome based on the coverage ratio of 2-kb windows. The scaffolds are ordered descendingly from the largest one on the x axis. Scaffolds are separated by black bars. (C) Comparison of the GC contents in C. eustigma and the evolutionarily related neutrophilic green algal species with sequence genomes. The x axis indicates the GC content, and the y axis indicates the proportion of the bin number divided by the total windows. We used 500-bp bins (with a 250-bp overlap) sliding along the genome. (D) Comparison of the number of genes in C. eustigma and C. reinhardtii whose functions were assigned to respective KEGG functional categories. Each bar indicates the number of genes that are assigned to the particular functional category. (E) Venn diagram of KEGG Orthology IDs to which one or more genes are assigned in C. eustigma and C. reinhardtii.
Most of the scaffolds consist of only single or duplicated sequences, with a few exceptions (for example, the largest scaffold is a chimera of single and duplicated regions) (Fig. 2B and Dataset S1). This result suggests that C. eustigma has experienced genomic duplication at the chromosomal level but not considerable rearrangements of the duplicated regions. Considering the length of the duplicated regions, the assembled genome size of C. eustigma was ∼110 Mb (Table S3). The difference between the estimated (∼130 Mb) and assembled (∼110 Mb) genome sizes is probably due to the difficulty in resolving repeats, which is often encountered in genome sequencing studies (17). The C. eustigma genome exhibits relatively low GC content (45%) compared with the genomes of other green algae (64% in C. reinhardtii; 56% in Volvox carteri; 67% in Chlorella variabilis; 53% in Coccomyxa subellipsoidea) (Fig. 2C).
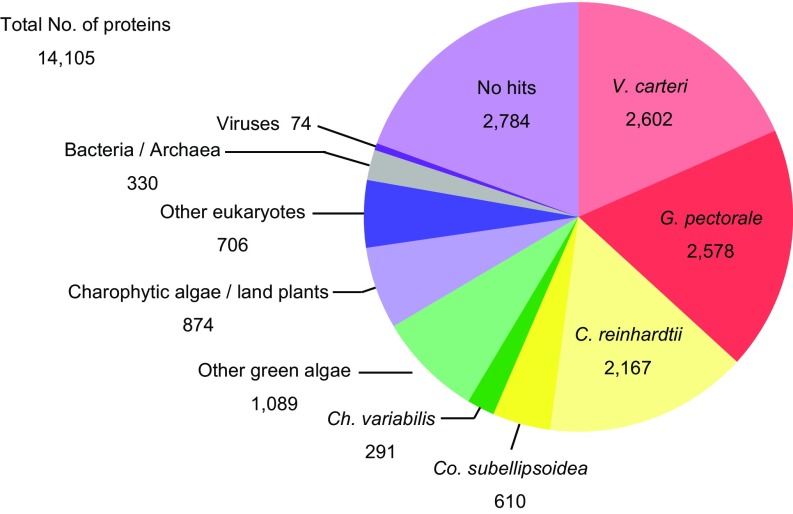
In the assembled C. eustigma draft genome, 14,105 protein-coding genes were identified by Augustus software with RNA-sequencing (RNA-seq) data (genes encoded in duplicated regions are counted as single genes) (Table S3). The BLASTP search against the Nationl Center for Biotechnology Information nonredundant (NCBI-nr) database (release 20160519) showed that 52.1% of C. eustigma proteins are most closely related to those of Volvocales (C. reinhardtii, Gonium pectorale, and V. carteri), whereas 19.7% showed no significant similarity to any known proteins (Fig. S1).
Fig. S1.
BLAST top-hit analysis of C. eustigma proteins against the NCBI-nr database. The pie chart shows the species distribution of the top BLASTP hits of the C. eustigma proteins (total 14,105 sequences). The BLASTP search was conducted with an e-value cutoff of 1e−5.
High Expression of HSP and PMA Genes in C. eustigma.
To compare the genomic contents between acidophile C. eustigma and neutrophile C. reinhardtii, functional annotations were assigned to C. eustigma and C. reinhardtii gene models. Predicted genes were assigned to the Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthology database through the KEGG Automatic Annotation Server (KAAS). The analysis assigned unique KEGG Orthology IDs to 4,470 C. eustigma and 4,741 C. reinhardtii protein-coding genes, respectively (Table S3). However, there were no marked differences in the number of genes classified into respective functional categories (Fig. 2D), and most of the KEGG Orthology IDs (3,006) were shared by the two species (Fig. 2E).
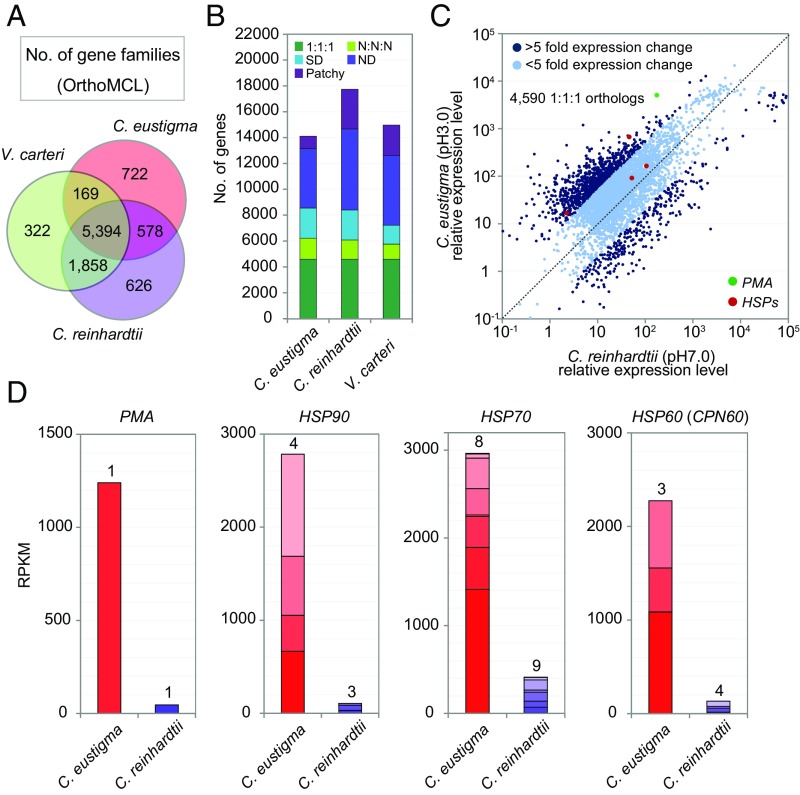
To examine the difference in the expression levels of the orthologous genes between C. eustigma and C. reinhardtii, we performed RNA-seq analyses of the two species under their individual optimal conditions (at 20 °C in the same autotrophic medium and at pH 3.0 for C. eustigma and pH 7.0 for C. reinhardtii). Before comparing the transcriptome, we identified 4,590 one-to-one orthologous genes in the three volvocalean species (C. eustigma, C. reinhardtii, and V. carteri) by OrthoMCL (Fig. 3 A and B). Of the 4,590 genes, 1,282 (∼30%) showed a greater than fivefold difference in the mRNA levels between C. eustigma and C. reinhardtii (Fig. 3C and Dataset S2). Notably, in the group that was up-regulated in C. eustigma, HSP genes were enriched (Fig. 3 C and D and Fig. S2). Consistent with the result at the mRNA level, previous studies showed that the acidophile C. acidophila (CCAP 11/137 isolated from acidic fresh water in Germany), which is closely related to C. eustigma (16), had higher basal HSP levels (HSP70, HSP60, and HSP20) than C. reinhardtii (18). These observations suggest that C. eustigma is constantly exposed to higher stress despite being adapted to an acidic environment.
Fig. 3.
Comparison of genome contents and transcriptome between the acidophile C. eustigma and neutrophile C. reinhardtii. (A) Venn diagram showing the number of protein families (by OrthoMCL) shared by C. eustigma, C. reinhardtii, and V. carteri genomes. (B) Gene orthologs of C. eustigma, C. reinhardtii, and V. carteri identified by OrthoMCL. “1:1:1” indicates an ortholog shared by three species as single copies; “N:N:N” indicates an ortholog shared by three species as multiple copies; “Patchy” indicates an ortholog shared by only two species. “ND” and “SD” indicate a species-specific gene in single or multiple copies, respectively. (C) Scatter plot of the mRNA levels of one-to-one orthologous genes between C. eustigma (pH 3.0, 20 °C) and C. reinhardtii (pH 7.0, 20 °C). The RPKM levels of 4,590 orthologous genes were plotted. (D) Comparison of RPKM levels of the PMA gene and HSP genes in C. eustigma (pH 3.0, 20 °C) and C. reinhardtii (pH 7.0, 20 °C). The number above the bar indicates the number of genes that exist in the respective nuclear genomes.
Fig. S2.
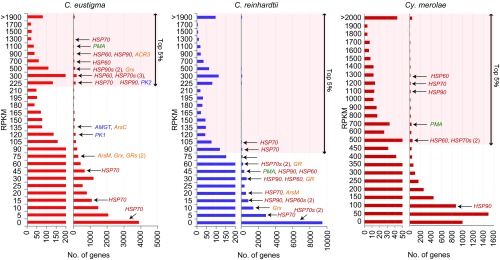
The relative levels of mRNAs in C. eustigma (pH 3.0, 20 °C), C. reinhardtii (pH 7.0, 20 °C), and the red alga Cy. merolae (pH 2.2, 42 °C). The left histograms show a magnified view of the right histograms. AMGT, amidinotransferase; GR, glutathione reductase; Grx, glutaredoxin; HSP, heat-shock protein; PK, phosphagen kinase; PMA, plasma membrane H+-ATPase.
In addition, we found that PMA was highly expressed in C. eustigma [151th highest reads per kilobase of transcript per million reads mapped (RPKM) value among 14,105 protein-coding genes] compared with C. reinhardtii (1,553th highest RPKM value among 17,741 protein-coding genes) (Fig. 3 C and D and Fig. S2). Maintenance of a neutral pH in the cytosol despite being in an acidic environment of pH 3 indicates the presence of a 104-fold proton gradient across the plasma membrane. It has been suggested that this proton gradient in acidophiles is achieved by a combination of active transport and low permeability of protons (19). In the acidophile Chlamydomonas sp. (ATCC PRA-125 isolated from acidic fresh water in Spain), it was previously shown that average cytosolic pH is maintained at pH 6.6 in the culture medium at both pH 2 and pH 7 (20). In C. reinhardtii cultured at pH 7, the average cytosolic pH was 7.1 (20). In addition, it was shown that 7% more ATP was consumed to remove protons entering the cytosol across the membrane at pH 2 than at pH 7 (20). Thus, the high expression of PMA in C. eustigma probably contributes to maintaining the high proton-pumping activity against the acidic environment.
Selective Loss of Acid-Producing Fermentation Pathways from C. eustigma.
The above comparison of genome contents showed that several hundred KEGG Orthology IDs (Fig. 2E) and gene families (Fig. 3A) are specific to either C. eustigma or C. reinhardtii, suggesting that the gene acquisitions and gene losses by C. eustigma after divergence from its neutrophilic ancestor also played roles in its adaptation to an acidic environment.
Regarding the gene losses by C. eustigma, we found that the genome had lost many genes involved in anaerobic fermentation pathways (Fig. 4 A and B). Several lineages of eukaryotic algae have evolved fermentation pathways that produce ATP when oxygenic respiration is compromised, for example, under anoxic/hypoxic conditions resulting from a low level of photosynthetic activity and local depletion of oxygen by microbial respiration (21). The alcohol fermentation pathway produces a diffusible, nonacidic, and relatively nontoxic end product, ethanol, while other pathways produce organic acids as end products that cause cytosolic acidification and damage (Fig. 4A) (22–24). Moreover, organic acids function as uncouplers of the respiratory chain at low pH by diffusion of the protonated form into the cell, followed by dissociation of a proton (19).
Fig. 4.
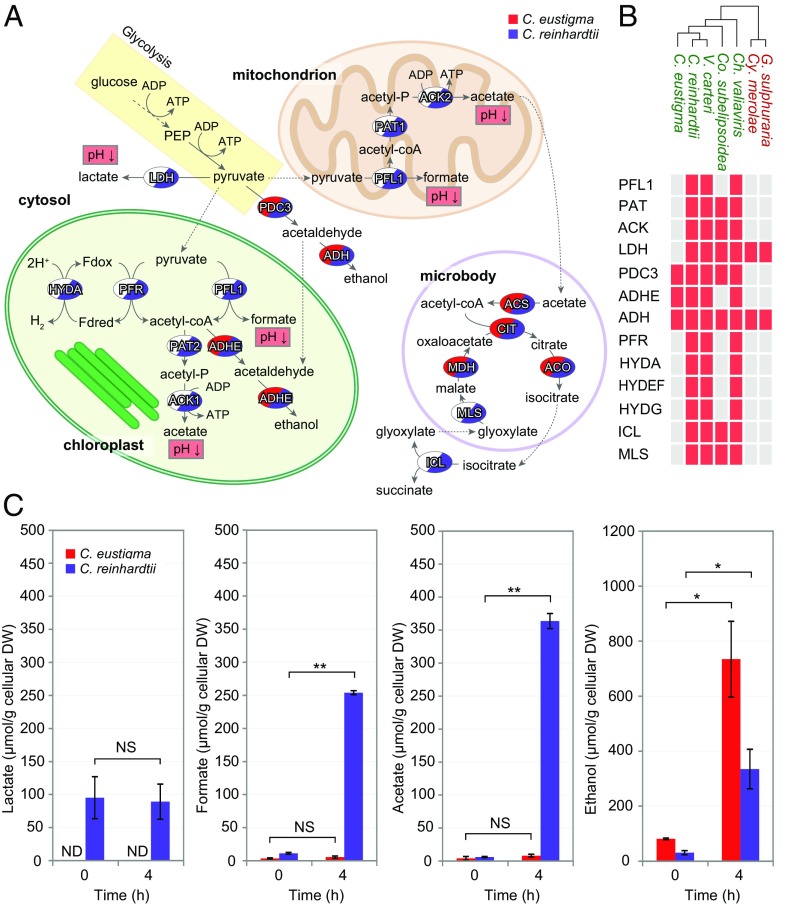
Loss of organic acid-producing fermentation pathways by C. eustigma. (A) Overview of the anaerobic fermentation pathways in eukaryotic algae (24, 66). Glucose (stored as starch) synthesized by photosynthesis is oxidized to pyruvate via glycolysis. Under anaerobic conditions, the conversion of pyruvate to acetyl-coA is catalyzed by the pyruvate formate lyase (PFL1) pathway that generates formate or by the pyruvate-ferredoxin oxidoreductase (PFR) and hydrogenase (HYDA) pathway that generates hydrogen. In C. reinhardtii, HYDEF and HYDG are essential to activate HYDA (67). Acetyl-CoA enters the phosphate acetyltransferase (PAT) and acetate kinase (ACK) pathway that generates acetate or the aldehyde/alcohol dehydrogenase (ADHE) pathway that generates ethanol. Pyruvate can also be used as a substrate to generate ethanol or lactate via the PDC3 (pyruvate decarboxylase 3) and alcohol dehydrogenase (ADH) pathway or lactate dehydrogenase (LDH) pathway, respectively. Acetate is used for lipid biosynthesis or is converted into acetyl-CoA by acetyl-CoA synthetase (ACS), which is further processed in the glyoxylate cycle to regenerate malate and succinate. ACO, aconitase; CIT, citrate synthase; ICL, isocitrate lyase; MDH, malate dehydrogenase; MLS, malate synthase; PEP, phosphoenolpyruvate. (B) Presence or absence of fermentation genes in the genomes of five green algae (shown in green) and two thermo-acidophilic red algae (shown in red). The red boxes indicate the presence of the gene, and white boxes indicate the absence of the gene. (C) Concentrations of lactate, formate, acetate, and ethanol in the algal culture medium before (0 h) and 4 h after the dark anaerobic treatment. The error bars represent the SD of three biological replicates. DW, dry weight; ND, not detected; NS, not statistically significant; *P < 0.02, **P < 0.01 (t test).
C. reinhardtii possesses fermentation pathways that produce both ethanol and organic acids, namely, lactate, formate, and acetate (Fig. 4 A and B) (24). Although the C. eustigma genome encodes pyruvate decarboxylase 3 (PDC3) and alcohol dehydrogenase (ADH) that produce ethanol, it lacks enzymes involved in organic acid fermentation pathways, such as lactate dehydrogenase (LDH) that produces lactate, pyruvate formate lyase (PFL) that produces formate, and both chloroplast and mitochondrial phosphate acetyltransferases (PAT2 and PAT1) and acetate kinases (ACK1 and ACK2) that produce acetate (Fig. 4 A and B). In addition to lacking these genes, C. eustigma lacks the genes encoding pyruvate:ferredoxin oxidoreductase (PFR) and hydrogenase (HYDA) and the proteins required for hydrogenase activation (HYDEF and HYDG) (25). All the above-mentioned genes absent in C. eustigma are present in other green algal genomes (C. reinhardtii, V. carteri, and Ch. Variabilis) (Fig. 4B and Table S4), suggesting that C. eustigma lost these genes during evolution after divergence from the common ancestor of C. reinhardtii and V. carteri. We found that some enzymes are also absent in Co. subellipsoidea C-169 (isolated from Antarctic dried algal peat) (26). However, based on the phylogenetic relationship between C. eustigma and Co. subellipsoidea, they probably lost these genes independently (Fig. 4B).
Table S4.
Genes involved in anaerobic energy metabolism in C. eustigma, C. reinhardtii, V. carteri, Co. subellipsoidea, Ch. variabilis, Cy. merolae, and G. sulphuraria
These genes have stop codons interrupting a coding sequence, resulting in a split into three fragments.
Consistent with the loss of genes involved in organic acid fermentation, HPLC analyses showed that C. eustigma produces little lactate, formate, and acetate (Fig. 4C). These three organic acids were detected in the supernatant fraction of C. reinhardtii culture but were scarcely detected in that of C. eustigma under aerobic conditions (Fig. 4C). When the cells were transferred to dark and anaerobic conditions, the formate and acetate levels increased in the supernatant fraction of C. reinhardtii culture 4 h after the transfer (Fig. 4C), as previously reported (27), but under these conditions these organic acids still were hardly detected in the supernatant fraction of C. eustigma (Fig. 4C). In contrast to the loss of organic acid fermentation by C. eustigma, a higher concentration of ethanol was detected in the supernatant fraction of C. eustigma culture than in that of C. reinhardtii by the gas chromatography analysis (Fig. 4C). The cellular ethanol level increased 4 h after the cells had been transferred from aerobic to dark and anaerobic conditions in both C. eustigma and C. reinhardtii (Fig. 4C). These results indicate that C. eustigma selectively lost organic acid-producing fermentative genes.
In the rice bean Vigna umbellata, it was reported that exposure of plants to low pH leads to the accumulation of formate. In addition, overexpression of V. umbellata formate dehydrogenase in tobacco resulted in decreased sensitivity to low pH and aluminum stresses by reducing the accumulation of formate (28). Thus, the loss of organic acid-producing fermentation pathways by C. eustigma probably contributed to the adaptation to acidic environments with high concentrations of metals. This probably also accounts for the independent loss of fermentation genes from Co. subellipsoidea because the genus Coccomyxa contains several acidophilic members (29, 30).
The genome analyses also showed that C. eustigma had lost the key enzymes of the glyoxylate cycle, namely, malate synthase (MLS) and isocitrate lyase (ICL) (Fig. 4 A and B). The glyoxylate cycle converts acetyl-CoA to succinate for the synthesis of carbohydrates and plays an essential role in cell growth on acetate (31). The loss of these enzymes is probably consistent with the fact that C. eustigma produces little acetate (Fig. 4C) and/or prevents cytosolic acidification that is caused by succinate production.
Acquisition of the Energy Shuttle and Buffering System Based on Amidinotransferase and Phosphagen Kinase by C. eustigma Through HGT.
Regarding the gene acquisition by C. eustigma, we found that the genome encodes two phosphagen kinases (PKs) and one amidinotransferase (AMGT), which were probably introduced through HGT (Fig. 5 A–C and Fig. S3). PK and AMGT exist in various animal, protozoan, and bacterial taxa and function as an energy shuttle and buffering system (32) (Fig. 5C). PK catalyzes the reversible transfer of a phosphate between ATP and guanidino compounds (e.g., arginine, creatine, glycocyamine, lombricine, and taurocyamine), which are produced from amino acids by AMGT (Fig. 5C) (32). However, PK or AMGT genes have not been identified in other Archaeplastida (land plants and eukaryotic algae whose chloroplasts are of cyanobacterial primary endosymbiotic origin) (11).
Fig. 5.
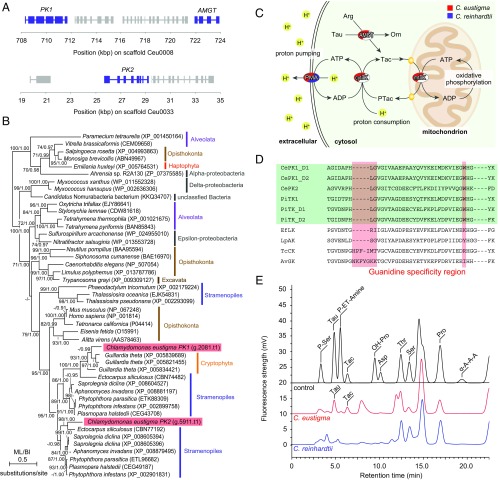
Acquisition of the PK–AMGT energy shuttle and buffering system by C. eustigma through HGT. (A) Genomic location of the two PK genes and one AMGT gene. PK1 and AMGT are encoded in the scaffold Ceu0008, and PK2 is encoded in the scaffold Ceu0033 in the C. eustigma genome. (B) Phylogenetic relationship of PK proteins. The tree was constructed by the ML method (RaxML 8.0.0). ML BP >50% obtained by RaxML, and BI >0.95 obtained by Bayesian analysis (MrBayes 3.2.6) are shown above the branches. The accession numbers of the sequences are shown along with the names of the species. The branch lengths reflect the evolutionary distances indicated by the scale bar. (C) Overview of the phosphagen kinase energy buffering system (32). Taurocyamine (Tac) is produced by amidinotransferase (AMGT) from l-arginine (Arg) and taurine (Tau). Tac is phosphorylated to produce phosphotaurocyamine (PTac) by mitochondrial taurocyamine kinase (mtTK) by consuming ATP produced by oxidative phosphorylation. PTac is dephosphorylated by cytosolic taurocyamine kinase (cytTK) to produce ATP. ATP is, for example, consumed by plasma membrane H+-ATPase (PMA) to pump protons from the cytosol to outside the cell. Orn, ornithine. (D) Amino acid sequence alignment of the guanidine specificity region in C. eustigma phosphagen kinase with those of other organisms. The guanidine specificity regions (33) are shaded red, and taurocyamine kinases are shaded green. D1 and D2 represent domain 1 and domain 2, respectively, of two-domain enzymes. AvGK, Alitta virens glycocyamine kinase; CePK, C. eustigma phosphagen kinase; EfLK, Eisenia fetida lombricine kinase; LpAK, Limulus polyphemus arginine kinase; PiTK, Phytophthora infestans taurocyamine kinase; TcCK, Tetronarce californica creatine kinase. (E) Fluorometric detection of amines in cellular extractions by HPLC/OPA. The control is a chromatogram of a standard mixture of amino acids and taurocyamine. Asp, aspartic acid; OH-Pro, hydroxyproline; P-ET-Amine, o-phosphoethanolamine; Pro, proline; P-Ser, o-phosphoserine; Ser, serine; Tac, taurocyamine; Tau, taurine; Thr, threonine; α-A-A-A, α-aminoadipic acid.
Fig. S3.
Phylogenetic relationship of AMGT proteins. The tree was constructed by the ML method (RaxML 8.0.0). ML BP >50% obtained by RaxML and BI >0.95 obtained by Bayesian analysis (MrBayes 3.2.6) are shown above the branches. The branch lengths reflect the evolutionary distances indicated by the scale bar. The accession numbers of the respective amino acid sequences are indicated with the species names.
In the C. eustigma genome, PK1 and AMGT are encoded in the same scaffold close to each other, whereas PK2 is encoded in another scaffold (Fig. 5A). Phylogenetic analysis showed that C. eustigma PK1 and PK2 are most closely related to PK proteins of cryptophytes and stramenopiles, respectively (Fig. 5B). In addition, C. eustigma AMGT is most closely related to that of bacteria and cryptophytes (Fig. S3). PK possesses a guanidine specificity region, which probably defines the substrate specificity (33). To determine the substrate of C. eustigma PKs, the guanidine specificity region was compared with PKs of other organisms for which substrates have been determined. In the amino acid sequence alignment, the guanidine specificity regions of C. eustigma PK1 and PK2 were found to be most closely related to taurocyamine kinase (TK) of Phytophthora infestans (Fig. 5D). By HPLC/o-phthalaldehyde (OPA) fluorometry, taurocyamine was detected in C. eustigma cellular extract but not in that of C. reinhardtii (Fig. 5E). These results suggest that C. eustigma acquired TK and AMGT as an l-arginine:taurine amidinotransferase through HGT.
The maintenance of a neutral cytosolic pH by acidophiles consumes a large amount of ATP, as described above (20). It was previously shown that the “artificial HGT of PK,” that is, the expression of exogenous arginine kinase in yeasts and Escherichia coli, which do not possess endogenous PKs, increased the resistance to transient pH reduction by building up an energy-storing phospho-arginine pool (34, 35). The RNA-seq results showed that PK1, PK2, and AMGT are relatively highly expressed in C. eustigma, exhibiting the 1,189th, 381th, and 1,050th highest RPKM values, respectively, among 14,105 protein-coding genes (Fig. S2 and Dataset S3). Thus, the acquisition of the PK–AMGT shuttle by C. eustigma has probably contributed to the supply of ATP needed to maintain cellular pH against an acidic environment (Fig. 5C).
Enhancement of Arsenic Biotransformation and Detoxification by C. eustigma Through HGT.
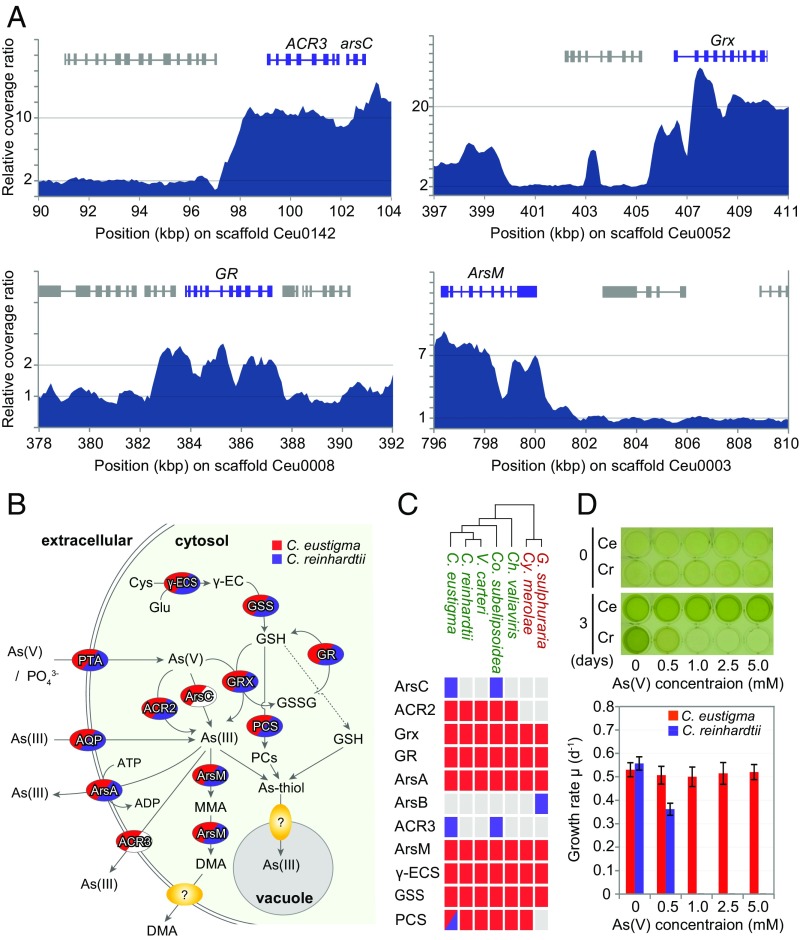
In addition to gene loss and acquisition through HGT, the genomic analysis of C. eustigma suggests that gene amplifications within the genome have also contributed to the adaptation to an acidic environment (Fig. 6A). It is known that natural acidic drainage often contains a very high concentration of toxic metals such as arsenic (36). In addition to accelerating metal solubilization, acidic water protonates arsenic, which accelerates the penetration of arsenic into cells (2). Arsenate (AsO43−), an analog of phosphate, is incorporated into cells along with phosphate, whereas arsenite (AsO33−) is incorporated into cells through aquaglycoporins (Fig. 6B) (37). Arsenite oxidizes thiols of biomolecules and causes strong oxidative stress (38). Consistent with the higher toxicity of arsenic in acidic environments, we found that C. eustigma tolerates a >10 times higher concentration of arsenate than C. reinhardtii (Fig. 6D). Genomic analyses showed that genes involved in arsenic biotransformation and detoxification (37) have been multiplied in the C. eustigma genome (Fig. 6A). The genome possesses approximately 10 copies of genes encoding arsenate reductase (ArsC) and arsenite efflux transporter (ACR3), which are located side-by-side in the genome, and approximately seven copies of the gene encoding arsenite S-adenosylmethionine methyltransferase (ArsM) (Fig. 6A). In addition, genes encoding glutaredoxin (Grx) (∼20 copies) and glutathione reductase (GR) (two copies), which are involved in the reduction of arsenate to arsenite (39), have also been multiplied in the genome (Fig. 6A). Consistent with the increase in the gene copy number, RNA-seq analysis showed that ArsM, Grx, and GR mRNA levels are higher in C. eustigma than in C. reinhardtii, even when both are cultured under their respective optimal growth conditions without arsenic (Fig. S2 and Datasets S3 and S4). In addition, ArsC and ACR3 are also relatively highly expressed in C. eustigma, exhibiting the 1,001th and 177th highest RPKM, respectively, among 14,105 protein-coding genes (Fig. S2 and Dataset S3).
Fig. 6.
Acquisition through HGT and expansion of genes in the arsenic biotransformation and detoxification system in C. eustigma. (A) Relative sequencing coverage ratio of Illumina MiSeq reads (200-bp window with 100-bp overlap) showing the copy numbers of arsenic biotransformation and detoxification genes. (B) Overview of the arsenic biotransformation and detoxification system (37). As(V) and As(III) are taken up through phosphate transporter (PTA) as a phosphate analog and through aquaglycoporins (AQP), respectively. As(V) is reduced to As(III) by glutaredoxin (Grx) by using glutathione (GSH) as a reductant or arsenate reductase ArsC or ACR2. As(III) is excreted by As(III)-specific transporter ArsA or ACR3. GSH is an antioxidant and is synthesized by γ-glutamylcysteine synthetase (γ-ECS) and glutathione synthetase (GSS). The methylation of As(III) to monomethylarsonic acid (MMA) and dimethylarsinic acid (DMA) is also thought to function as inorganic arsenic biotransformation and detoxification. As(III) is chelated by phytochelatins (PCs), which are synthesized by phytochelatin synthase (PCS) or GSH to form the As(III)-thiol complex, which is finally transported into the vacuole. (C) Presence or absence of arsenic biotransformation and detoxification genes in the genomes of five green algae (shown in green) and two thermo-acidophilic red algae (shown in red). The red and blue boxes indicate the presence of the gene; blue boxes indicate a gene that was horizontally transferred. The gray boxes indicate the absence of the gene. (D) Effects of the concentration of arsenate As(V) on the growth of C. eustigma and C. reinhardtii. Cells were cultured in modified original medium containing both 1 mM phosphate (originally 10 mM) and various concentrations of arsenate for 3 d. The error bars in the graph represent the SD of three biological replicates.
Several studies have already succeeded in enhancing the tolerance to arsenic by artificial HGT, for example, by overexpressing E. coli ArsC and γ-glutamylcysteine synthase (40) (to increase the thiol pool) or overexpression of the yeast ACR3 in Arabidopsis thaliana (41). Thus, the multiplication and high expression of arsenic biotransformation and detoxification genes in C. eustigma have probably contributed to the high algal resistance to arsenic.
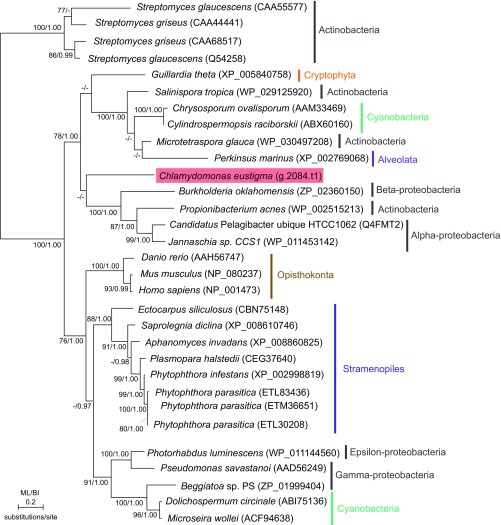
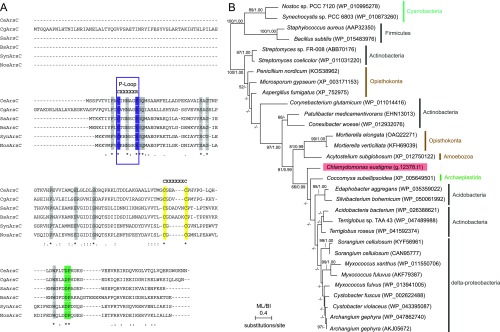
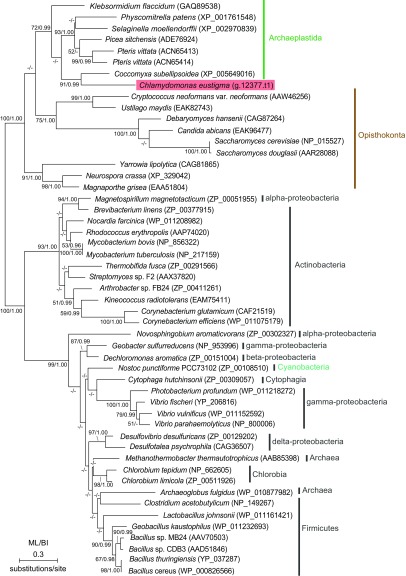
The comparison of green algal genomes showed that, among the proteins related to arsenic biotransformation and detoxification, ArsC and ACR3 are not encoded in other green algal genomes except for those of C. eustigma and Co. subellipsoidea (Fig. 6C and Table S5). Based on the phylogenetic relationship between these two species, C. eustigma and Co. subellipsoidea probably acquired ArsC and ACR3 genes independently (Fig. 6C). In the phylogenetic analyses, ArsC of C. eustigma and C. subellipsoidea formed a clade with those of acidobacteria, actinobacteria, and δ-proteobacteria (Fig. S4), suggesting the bacterial HGT origin of C. eustigma ArsC. On the other hand, C. eustigma and Co. subellipsoidea ACR3 formed a clade with proteins of charophycean algae and certain land plant species, and this clade is a sister group of fungal proteins (Fig. S5). Thus, the origin of C. eustigma ACR3 is not clear at this point; however, given that only a limited number of green algae and land plants possess ACR3 (Fig. S5), it is likely that ACR3 was acquired by these species multiple times independently through HGT. Thus, the multiplication of both genes derived from their eukaryotic ancestor (ACR3, Grx, and GR) and genes acquired through HGT (ArsC and ACR3) probably contributed to the adaptation of C. eustigma.
Table S5.
Genes involved in arsenic biotransformation and detoxification in C. eustigma, C. reinhardtii, V. carteri, Co. subellipsoidea, Ch. variabilis, Cy. merolae, and G. sulphuraria
Fig. S4.
Multiple sequence alignment and phylogenetic relationship of ArsC proteins. (A) Multiple sequence alignment of the C. eustigma ArsC protein with bacterial arsenate reductase. Proteins of Corynebacterium glutamicum (WP_011014416), Staphylococcus aureus (AAP32350), Bacillus subtilis (WP_015483976), Synechocystis sp. PCC6803 (WP_010873260), and Nostoc sp. PCC 7120 (WP_010995278) were included. The active site motif, Cys-XXXXX-Arg (the P-loop, an anion binding motif) (82), is boxed in purple. The two distal cysteine residues that participate in the catalytic disulfide bond cascade (82) are shaded yellow. The conserved Asp-Pro sequence, which is important for catalysis (82), is shaded green. Other conserved residues are shaded gray. (B) Phylogenetic relationship of ArsC proteins. The tree was constructed by the ML method (RaxML 8.0.0). ML BP >50% obtained by RaxML and BI >0.95 obtained by Bayesian analysis (MrBayes 3.2.6) are shown above the branches. The branch lengths reflect the evolutionary distances indicated by the scale bar. The accession numbers of the respective amino acid sequences are indicated with the species names.
Fig. S5.
Phylogenetic relationship of ACR3 proteins. The tree was constructed by the ML method (RaxML 8.0.0). ML BP >50% obtained by RaxML and BI >0.95 obtained by Bayesian analysis (MrBayes 3.2.6) are shown above the branches. The branch lengths reflect the evolutionary distances indicated by the scale bar. The accession numbers of the respective amino acid sequences are indicated with the species names.
Discussion
The above analyses showed that the C. eustigma genome has experienced large-scale duplication throughout its genome (Fig. 2B) and has a lower GC content than evolutionarily related neutrophilic green algae sequenced thus far (Fig. 2C). However, it is currently unclear whether there are any relationships between these features in the genome structure and the adaptation to an acidic environment. Generally, genome or gene duplication is widely considered to facilitate environmental adaptation because the redundancy generated allows the evolution of new beneficial gene functions that are otherwise prohibited due to functional constraints (42). Genomic GC content is predicted to affect genome functioning and species ecology significantly. However, the biological significance of GC content diversity remains elusive because of a lack of sufficiently robust genomic data (43).
Comparative genome and transcriptome analyses suggest that the following features of genomic evolution have contributed to the adaptation of C. eustigma to an acidic environment. (i) HSPs and PMA became expressed at high levels. (ii) The genome lost fermentative genes that produce organic acids in the cell. (iii) The genome acquired genes encoding the PK–AMGT energy shuttle and buffering system and genes that are involved in arsenic biotransformation and detoxification through HGT. (iv) The genes involved in arsenic biotransformation and detoxification, derived from a green algal ancestor or acquired through HGT, were multiplied in the genome.
In addition, based on this study and the results of previous studies in other acidophilic algae, it is suggested that these genomic changes are probably common trends in the adaptation to acidic or other extreme environments, as discussed below. Regarding (i), we also found that HSPs are highly expressed in the thermo-acidophilic red alga Cy. merolae under its optimal conditions (in an autotrophic medium at pH 2.5 and 42 °C), as in the case of C. eustigma compared with C. reinhardtii (Fig. S2 and Dataset S5). Regarding (ii), Co. subellipsoidea also lost organic acid-producing fermentation genes independently from C. eustigma (Fig. 4 A and B). The genus Coccomyxa contains several acidophiles (29, 30). However, it is currently unclear whether there is a correlation between the loss of organic acid-producing fermentation and adaptation to an acidic environment in acidophilic red algae, because they possess only the lactate fermentation pathway and alcohol dehydrogenases (Fig. 4 A and B). Regarding (iii), although the HGT of PK–AMGT into acidophiles has not been reported in other acidophiles, the acquisition of arsenic biotransformation and detoxification genes through HGT has been found in the green alga Co. subellipsoidea and thermo-acidophilic red algae (Fig. 6C) (8). The green alga Co. subellipsoidea acquired ACR3 (Fig. 6C and Fig. S5), and the red alga G. sulphuraria acquired ArsB through HGT (8) (Fig. 6C). Regarding (iv), in the G. sulphuraria genome, genes of the chloride channel and chloride carrier/channel families have been multiplied and are thought to be important to acid tolerance (8). In addition, an archaeal ATPase of HGT origin has been multiplied and probably contributes to heat tolerance (8).
This study and recent studies on the genomes of acidophiles have started to reveal commonalities in genomic evolution regarding adaptation to an acidic environment. Besides increasing our understanding of evolution, this information could also have important applications. Microalgae have been cultivated at a large scale to produce functional foods and pigments and are also considered to be an alternative source for biofuels because of their relatively rapid growth to a high concentration (44). Acidophilic microalgae have an advantage in that they can be cultivated outdoors without the risk of contamination by other undesirable organisms (45). In addition, trials using acidophiles for bioremediation (46) and metal recovery have also been initiated (47). An understanding of the genetic basis of acid and/or metal tolerance is also necessary to confer such abilities to other organisms.
Methods
Algal Strains.
C. eustigma (NIES-2499; Microbial Culture Collection at the National Institute for Environmental Studies) and C. reinhardtii 137c mt+ were used in the current study. C. eustigma and C. reinhardtii were maintained with gyration (100 rpm) on a rotary shaker (NR-2; Taitec) in photoautotrophic medium (9.35 mM NH4Cl, 81.15 μM MgSO4·7H2O, 68.04 μM CaCl2, 10 mM KH2PO4/K2HPO4, 59.2 μM FeCl3, 0.73 μM MnCl2·4H2O, 0.31 μM ZnSO4·7H2O, 0.0672 μM CoCl2·6H2O, 0.0413 μM Na2MoO4·2H2O, 0.5046 μM CuSO4·5H2O, 13.75 μM Na2EDTA·2H2O) at pH 3.0 and pH 7.0, respectively, at 21 °C under a 12-h light/12-h dark photoperiod (30 μE⋅m−2⋅s−1).
Genomic DNA Sequencing.
Genomic DNA of C. eustigma was extracted, and sequencing libraries were prepared according to ref. 48. The shotgun and paired-end libraries (8 kb) were sequenced by Roche 454 GS FLX+ Titanium (Roche Diagnostics). The paired-end (400-bp) library was sequenced by HiSeq 2500 with 100 base-paired end format with the TruSeq SBS kit v3 (Illumina, Inc.). The paired-end (800-bp) and mate-pair libraries (3, 5, and 8 kb) were sequenced by MiSeq (Illumina, Inc.) with the MiSeq reagent kit version 3 (600 cycles; Illumina). The MiSeq reads were filtered using ShortReadManager (49), based on a 17-mer frequency.
Estimation of the Genome Size by K-mer Analysis.
The MiSeq paired-end reads were used for the K-mer 31 frequency distribution analysis using JELLYFISH (50). The total genome size was estimated by analyzing the occurrence and distribution of K-mers using the following formula: Estimated genome size in base pairs = K-mer number/depth. The 31 K-mer depth distribution of the MiSeq paired-end reads exhibited two peaks (Fig. 2A). The estimated genome size of C. eustigma was ∼130 Mb, when the ×19 was considered as the main peak.
Genome Assembly, Scaffolding, and Gap Closing.
The Roche 454 shotgun and paired-end reads were assembled de novo by Newbler version 2.9 (Roche) with the following parameters: -mi 98 -mL 80 -scaffold -large -s 500. Subsequent scaffolding of the Newbler output contigs was performed by SSPACE (51) using the Illumina paired-end and mate-pair information (Table S2). GMcloser (52) was used for gap filling with preassembled contigs and Illumina paired-end reads. The genome sequence was improved with Illumina paired-end reads using iCORN2 (53). To remove mitochondrial and chloroplast DNA sequences, tblastn (54) searches were performed against scaffolds by using amino acid sequences of C. reinhardtii mitochondrion- and chloroplast-encoded proteins as queries. By the tblastn search, one mitochondrial DNA scaffold and two chloroplast DNA scaffolds were identified, and these scaffolds were removed from the assembly.
Estimation of Coverage Ratio in the C. eustigma Genome.
The sequencing coverage ratio was assessed by calculating normalized coverage depth followed by manual inspection of depth variation along each scaffold. The MiSeq read data (6,660,746 reads) were mapped to the scaffolds using Bowtie2 versiom 2.1.0 (55) with default settings. Mapped reads in sequence alignment/map (SAM) format were converted to the binary version of the SAM file (BAM format) using the Samtools version 0.1.19 (56) <view>, <sort>, and <index> commands. The aligned reads in BAM format were filtered for duplicates using the Samtools version 0.1.19 <rmdup> command. After the removal of duplicates, BAM files were converted to browser extensible data (BED) format using the Bedtools version 2.17 (57) <bamToBed> command. Genome-wide windows were defined using the Bedtools <makewindows> command, and then coverage depth of each individual window was calculated using the Bedtools <coverage> command. The histogram of the coverage ratio exhibited two major peaks that probably correspond to single and duplicated regions. The relative coverage ratio (shown in Figs. 2B and 6A) was normalized by the averaged coverage depth of the probable single regions.
Prediction and Annotation of the Nuclear Genes.
Nuclear genes were predicted by Augustus 3.0.3 (58). Assembled transcript sequences were mapped to the scaffolds by BLAT (59) to assess the likelihood that each sequence was indeed a transcript. The manually curated 1,900 gene models were used as Augustus training sets, and 14,105 genes were predicted by Augustus with transcript evidence. The KEGG Orthology ID assignment was performed for all predicted genes in the C. eustigma and C. reinhardtii genomes. The assignments were performed by the KAAS (60).
Comparison of mRNA Levels of Orthologous Genes in C. eustigma and C. reinhardtii.
To identify one-to-one orthologous genes in the three Volvocales (C. eustigma, C. reinhardtii, and V. carteri), gene clustering analysis was performed by OrthoMCL (61) with the following parameters: inflation value = 1.5, percentMatchCutoff = 50, and evalueExponentCutoff = −10. A BLASTP search for predicted amino acid sequences with an E-value of 1e−10 in the three algal species was performed using NCBI BLAST+ version 2.2.30. In total, we found 4,590 one-to-one ortholog pairs. Gene-expression scores were obtained from RNA-seq data by mapping the clean reads to the genes by Bowtie2 version 2.1.0 (55). SAMtools (56), BEDtools (57), and R version 2.14.2 (62) were used to calculate the tag-count data that were mapped to the coding genes. Normalization of the orthologous gene-expression scores was performed by RPKM normalization. After obtaining the normalized expression scores of orthologous genes for each sample, the scores were log (base 10)-transformed and plotted to produce a scatter graph for comparison of the expression scores of the two algal species.
Comparison of mRNA Levels in Algal Species.
The RNA-seq reads were mapped to the C. eustigma, C. reinhardtii (JGI, version 5.5), or Cy. merolae (Cyanidioschyzon merolae Genome Project) coding sequences by Bowtie2 (55) with the default parameters. The Bowtie2 outputs were processed to obtain tag counts. Since it has been shown that the GC content affects the read abundances in an RNA-seq dataset (63), counts were full-quantile normalized within a sample by GC content bias-correction methods implemented in the EDASeq R package (64). These normalized counts were used to calculate the mRNA level of each gene (in RPKM units) in the algal samples according to ref. 65.
Data Availability.
The C. eustigma NIES-2499 whole-genome and gene models have been deposited in DNA Data Bank of Japan (DDBJ)/European Molecular Biology Laboratory (EMBL)/GenBank under the accession code PRJDB5468. The dataset includes sequences of the nuclear and mitochondrial genomes. Because the chloroplast genome was highly repetitive and the genome could not be assembled well, the chloroplast genome was omitted from the dataset. The RNA-seq data of C. eustigma and C. reinhardtii have been deposited in DDBJ/EMBL/GenBank (accession codes PRJDB6154 and PRJDB6155, respectively).
SI Methods
Determination of Optimal pH Conditions.
To determine the optimal pH for C. eustigma and C. reinhardtii, cells were cultured at 20 °C in photoautotrophic medium buffered at eight different pH values (pH 1.0–8.0). The medium was buffered with 20 mM of the chemicals indicated below, except for the pH 1.0, 2.0, and 3.0 media (the pH levels of these media were adjusted with 1 N H2SO4/1 N KOH). The chemicals used to buffer the pH were 3,3-dimethylglutaric acid (DMGA) for pH 4.0, 2-N-morpholino)ethanesulfonic acid (Mes) for pH 5.0, piperazine-N,N′-bis (Pipes) for pH 6.0, 3-N-morpholino)propanesulfonic acid (Mops) for pH 7.0, and Hepes for pH 8.0. C. eustigma and C. reinhardtii were cultured at pH 3.0 and pH 7.0, respectively, until they reached an OD750 of 1.0–2.0. Then, the cells were harvested by centrifugation at 1,500 × g for 5 min and were gently resuspended into 50 mL of each medium (from pH 1.0 to pH 8.0) to give an OD750 of 0.5. Cells were cultured at 20 °C in 100-mL test tubes (∼3 cm thick) under continuous light (90 µE⋅m−2⋅s−1) with aeration (0.3 L ambient air/min) for 24 h. OD750 was measured with a spectrophotometer (SmartSpec Plus; Bio-Rad). Growth rates were determined according to the method described in ref. 68.
Reidentification of C. eustigma in the AMD and Quality Analyses of the AMD Water.
C. eustigma (NIES-2499) was originally isolated together with acidophilic mosses from AMD of an abandoned sulfur mine in Nagano Prefecture, Japan (14). In September 2013, we visited the same AMD (pH 2.13, 14.5 °C), which is located in Joshinetsu-kogen National Park, Japan. We harvested the mosses from the AMD with administrators of Hokushin Forest Office, which is responsible for management of the forest. The moss samples, to which algae adhere, were incubated in M-Allen liquid medium (69) at pH 2.0 and 15 °C under continuous-light conditions (20 µE⋅m2⋅s−1). The algae in the liquid culture were serially diluted and spread on M-Allen agar medium. After incubation at 20 °C under continuous-light conditions (20 µE⋅m2⋅s−1), green colonies appeared. As determined by microscopy, the cells in most of these colonies exhibited Chlamydomonas-like morphology. The 18S rDNA of the alga was also amplified by PCR and sequenced as described previously (70). The ∼1.5-kb sequence of 18S rDNA of the isolated alga was completely identical to that of C. eustigma (NIES-2499).
The nutrient contents of the AMD water were determined according to ref. 71 as follows. Fe2+ and Fe3+ were quantified by the 1,10-phenanthroline photometric method. Al3+, Cu2+, and Zn2+ were quantified by inductively coupled plasma mass spectrometry. Mn2+, Na+, K+, Mg2+, and Ca2+ were quantified by atomic absorption spectrometry. SO42− was quantified by the barium sulfate turbidimetric method. Cl− was quantified by Mohr’s method. NH4+ was quantified by the indophenol method. PO43− was quantified by the molybdenum blue colorimetric method. H2SiO3 was quantified by the molybdenum yellow colorimetric method.
Characterization of Arsenate Tolerance.
The comparison of arsenate tolerance was determined as described in ref. 72, with minor modifications. C. eustigma and C. reinhardtii cells cultured in the medium at pH 3.0 and at pH 7.0, respectively, and 20 °C (OD750 of 1.0–2.0) were harvested by centrifugation at 1,500 × g for 5 min and then were gently resuspended in the same fresh medium supplemented with 1 mM phosphate and a series of concentrations of potassium arsenate (KH2AsO4), as indicated in Fig. 6D, to give an OD750 of 0.5. Cells were cultured with gyration in a 24-well culture plate at 20 °C under continuous light (100⋅μE⋅m−2⋅s−1) for 3 d.
RNA-Seq Analyses.
C. eustigma and C. reinhardtii were cultured in 50 mL of photoautotrophic medium at pH 3.0 and pH 7.0, respectively, in 100-mL test tubes (∼3 cm thick) at 20 °C under continuous-light conditions (100 μE⋅m−2⋅s−1) with aeration (0.3 L ambient air/min). Total RNA was extracted from 10 mL of log-phase culture (∼1.0 × 106 cells/mL) of each of C. eustigma and C. reinhardtii. The unicellular red alga Cy. merolae was cultured in 50 mL of 2× Allen’s medium (a photoautotrophic medium) at pH 2.5 in 100-mL test tubes (∼3 cm thick) and 42 °C under continuous-light conditions (100 μE⋅m−2⋅s−1) with aeration (0.3 L ambient air/min). Total RNA was extracted from 20 mL of log-phase culture (∼1.0 × 107 cells/mL) of Cy. merolae. Cell numbers were determined using an improved Neubauer hemacytometer.
mRNA of respective samples was purified from 10 μg of total RNA with Dynabeads Oligo(dT)25 (Life Technologies), according to ref. 73. Sequencing libraries were prepared according to ref. 74. Sequencing was performed by HiSeq 2500 with 100 base-paired end format with TruSeq SBS kit v3 (Illumina). The reads were cleaned up using the cutadapt program version 1.81 (75) by trimming low-quality ends (<QV30) and adapter sequences and by discarding reads shorter than 50 bp.
Molecular Phylogenetic Analyses.
The nucleic acid (23S rDNA, 16S rDNA, atpB, psaA, psaB, psbC, and rbcL) or amino acid (TK, AMGT, ArsC, and ACR3 proteins) sequences were aligned using MAFFT version 7.212 (76), and ambiguous sites were excluded by Gblocks version 0.91b (77). Concatenation of the datasets (chloroplast 16S rDNA, 23S rDNA, atpB, psaA, psaB, psbC, and rbcL) was performed with Kakusan4 (78). Substitution models were selected by Kakusan4 or Aminosan (78) (GTR+G for 16S rDNA, 23S rDNA, atpB, psaA, psaB, psbC, and rbcL; LG+G for TK, AMGT, and ArsC proteins; LG+F+G for ACR3 protein). The ML analysis and 1,000 pseudoreplicates of bootstrap analyses were performed by RaxML 8.0.0 (79). Bayesian analyses were performed by MrBayes 3.2.6 (80) using the optimal substitution models with 1 million iterations, a subsample frequency of 1,000, and a burn-in of 100,000. The ML and Bayesian analyses of the concatenated datasets (21 taxa, 11,367 sites) were calculated under separate model conditions.
Quantification of Extracellular Metabolites by HPLC/Gas Chromatography.
C. eustigma and C. reinhardtii cells cultured in the media at pH 3.0 and pH 7.0, respectively, and 20 °C (OD750 of 1.0–2.0) were harvested by centrifugation at 1,500 × g for 5 min and then were gently resuspended into 50 mL of respective fresh medium to give an OD750 of 0.5. These cells were then cultured at 20 °C under continuous light (100 μE⋅m2⋅s−1) with aeration (0.25 L ambient air/min) for 2 d and then were continuously flushed with N2 gas in the dark. C. eustigma and C. reinhardtii cells were harvested by centrifugation at 1,500 × g for 5 min from respective 10-mL cultures just before and 4 h after the onset of aeration with N2 in the dark. The supernatant was transferred to a new vial, and the sample was then frozen in liquid N2 for subsequent analyses. Cellular dry weights were measured according to the method described in ref. 68. Organic acid analysis was performed by HPLC (Shimadzu Prominence; Shimadzu). The supernatant sample was thawed, centrifuged, and filtered (pore size of 0.22 μm), and then 50 μL of the sample was injected using an autosampler (SIL-20AC; Shimadzu). The sample was separated with two serial ion-exclusion chromatography columns (Shim-pack SCR-102H; Shimadzu) equipped with a guard column (Shim-pack SCR-102HG; Shimadzu) at 45 °C in a mobile phase of 5 mM p-toluenesulfonic acid, and then the eluent pH was adjusted with a buffering solution containing 5 mM p-toluenesulfonic acid, 20 mM Bis-Tris, and 0.1 mM EDTA at a flow rate of 0.8 mL/min. Each acid was quantified with a conductivity detector (CDD-10AVP; Shimadzu). Retention peaks for the various organic acids were recorded by LabSolutions LC/gas chromatography software, and quantification was performed by comparisons with the absorption of known amounts of a standard for respective organic acids. Ethanol analysis was performed by gas chromatography with an FID detector (GC-2014; Shimadzu) using a glass column of Thermon-1000 5% Sunpak-A (Shimadzu). The column temperature was maintained at 130 °C, and the temperatures of the injector and detector were maintained at 180 °C. N2 was used as a carrier gas at a flow rate of 50 mL/min. Aliquots (1.0 μL) of samples were injected using an autosampler (AOC-20c; Shimadzu).
Detection of Taurocyamine by HPLC/OPA Fluorometry.
C. eustigma and C. reinhardtii cells cultured in medium at pH 3.0 and pH 7.0, respectively, and 20 °C (OD750 of ∼1.0) were harvested by centrifugation at 1,500 × g for 5 min and then were gently resuspended into 400 mL of respective fresh medium to give an OD750 of 0.5. Cells were cultured in 700-mL flat bottles (∼5 cm thick) at 20 °C under continuous-light conditions (100 μE⋅m−2⋅s−1) with aeration (1.0 L ambient air/min) for 1 d. They were then harvested from the respective 50-mL cultures by centrifugation at 1,500 × g for 5 min and were stored at −80 °C until use. Taurocyamine was extracted from the cells as described by ref. 81, with minor modifications. Briefly, taurocyamine was extracted from the cell pellets twice with 1 mL of ice-cold 80% ethanol and then was centrifuged (16,100 × g for 5 min at 4 °C). The supernatants (2 mL) were extracted with 2 mL of ice-cold chloroform to remove pigments and fatty materials. The aqueous fraction was dried by a vacuum freeze dryer (EYELA, FRD-1200; Rikakikai Co. Ltd.). The dried sample was then dissolved in 100 µL of sodium citrate buffer (pH 2.2) solution (Wako Pure Chemical Industries). Analysis of amines was performed by HPLC (Shimadzu Prominence; Shimadzu) with postcolumn fluorescence derivatization using OPA/N-acetylcysteine. Then, 10-μL aliquots of the samples were injected using an autosampler (SIL-20AC; Shimadzu). Samples were separated with an ion-exclusion chromatography column (Shim-pack Amino-Na; Shimadzu) using the Amino Acid Mobile Phase Kit (Na Type; Shimadzu) with gradient elution according to the manufacturer’s protocol (flow rate, 0.4 mL/min; column temperature, 60 °C). A trap column (ISC-30/S0504 Na; Shimadzu) was placed between the pump and the autosampler to trap ammonia. The postcolumn fluorescence derivatization was conducted with an Amino Acid Reaction Reagent Kit (Shimadzu) according to the manufacturer’s protocol (flow rate, 0.2 mL/min; reaction temperature, 60 °C). Derivatized amines were monitored by a fluorescence detector (RF-20Axs; Shimadzu) with 350-nm excitation and 450-nm emission. Retention peaks for the various amines were recorded by LabSolutions LC/gas chromatography software.
Supplementary Material
Acknowledgments
We thank Dr. T. Kuroiwa, Dr. H. Kuroiwa, Dr. K. Tanaka, and Dr. Y. Kabeya for kind encouragement and advice on this work; Dr. K. Hori, Dr. N. V. Sasaki, Dr. M. Ishikawa, Dr. T. Fujisawa, and Mr. Y. Kazama for advice on bioinformatic analyses; Dr. U. Goodenough and Dr. Y. Nishimura for advice on Chlamydomonas research; Mr. T. Sone for advice on RNA extraction; and members of the S.-y.M. Laboratory for their technical advice and support. This work was supported by the Core Research for Evolutional Science and Technology Program of the Japan Science and Technology Agency (S.-y.M.), by Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science 25251039 (to S.-y.M.), and by the Ministry of Education, Culture, Sports, Science and Technology-supported Program for Strategic Research Foundation at Private Universities 2013–2017 S1311017. Computations were partially performed on the National Institute of Genetics (NIG) supercomputer at the Research Organization of Information and Systems of the NIG.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in DNA Data Bank of Japan/European Molecular Biology Laboratory-European Bioinformatics Institute/GenBank under the accession codes PRJDB5468, PRJDB6154, and PRJDB6155.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707072114/-/DCSupplemental.
References
- 1.Oren A. Encyclopedia of Life Sciences. Wiley; Chichester, UK: 2010. Acidophiles; pp. 192–206. [Google Scholar]
- 2.Gross W. Ecophysiology of algae living in highly acidic environments. Hydrobiologia. 2000;33:31–37. [Google Scholar]
- 3.Ferris MJ, et al. Algal species and light microenvironment in a low-pH, geothermal microbial mat community. Appl Environ Microbiol. 2005;71:7164–7171. doi: 10.1128/AEM.71.11.7164-7171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Novis P, Harding JS. Extreme acidophiles: Freshwater algae associated with acid mine drainage. In: Seckbach J, editor. Algae and Cyanobacteria in Extreme Environments. Springer; Heidelberg: 2007. pp. 443–463. [Google Scholar]
- 5.Pedrozo F, et al. First results on the water chemistry, algae and trophic status of an Andean acidic lake system of volcanic origin in Patagonia (Lake Caviahue) Hydrobiologia. 2001;452:129–137. [Google Scholar]
- 6.Amaral Zettler LA, et al. Microbiology: Eukaryotic diversity in Spain’s River of Fire. Nature. 2002;417:137. doi: 10.1038/417137a. [DOI] [PubMed] [Google Scholar]
- 7.Matsuzaki M, et al. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature. 2004;428:653–657. doi: 10.1038/nature02398. [DOI] [PubMed] [Google Scholar]
- 8.Schönknecht G, et al. Gene transfer from bacteria and archaea facilitated evolution of an extremophilic eukaryote. Science. 2013;339:1207–1210. doi: 10.1126/science.1231707. [DOI] [PubMed] [Google Scholar]
- 9.Qiu H, et al. Adaptation through horizontal gene transfer in the cryptoendolithic red alga Galdieria phlegrea. Curr Biol. 2013;23:R865–R866. doi: 10.1016/j.cub.2013.08.046. [DOI] [PubMed] [Google Scholar]
- 10.Olsson S, et al. Horizontal gene transfer of phytochelatin synthases from bacteria to extremophilic green algae. Microb Ecol. 2017;73:50–60. doi: 10.1007/s00248-016-0848-z. [DOI] [PubMed] [Google Scholar]
- 11.Adl SM, et al. The revised classification of eukaryotes. J Eukaryot Microbiol. 2012;59:429–493, and erratum (2013) 60:321. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoon HS, Hackett JD, Pinto G, Bhattacharya D. The single, ancient origin of chromist plastids. Proc Natl Acad Sci USA. 2002;99:15507–15512. doi: 10.1073/pnas.242379899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merchant SS, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higuchi S, et al. Morphology and phylogenetic position of a mat-forming green plant from acidic rivers in Japan. J Plant Res. 2003;116:461–467. doi: 10.1007/s10265-003-0125-3. [DOI] [PubMed] [Google Scholar]
- 15.Nordstrom DK, Alpers CN. Negative pH, efflorescent mineralogy, and consequences for environmental restoration at the Iron Mountain Superfund site, California. Proc Natl Acad Sci USA. 1999;96:3455–3462. doi: 10.1073/pnas.96.7.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yumoto K, Kasai F, Kawachi M. Taxonomic re-examination of Chlamydomonas strains maintained in the NIES-collection. Microbiol Cult Collect. 2013;29:1–12. [Google Scholar]
- 17.Schatz MC, Delcher AL, Salzberg SL. Assembly of large genomes using second-generation sequencing. Genome Res. 2010;20:1165–1173. doi: 10.1101/gr.101360.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerloff-Elias A, Barua D, Mölich A, Spijkerman E. Temperature- and pH-dependent accumulation of heat-shock proteins in the acidophilic green alga Chlamydomonas acidophila. FEMS Microbiol Ecol. 2006;56:345–354. doi: 10.1111/j.1574-6941.2006.00078.x. [DOI] [PubMed] [Google Scholar]
- 19.Baker-Austin C, Dopson M. Life in acid: pH homeostasis in acidophiles. Trends Microbiol. 2007;15:165–171. doi: 10.1016/j.tim.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 20.Messerli MA, et al. Life at acidic pH imposes an increased energetic cost for a eukaryotic acidophile. J Exp Biol. 2005;208:2569–2579. doi: 10.1242/jeb.01660. [DOI] [PubMed] [Google Scholar]
- 21.Maier RM, Pepper IL, Gerba CP. Environmental Microbiology. Gulf Professional Publishing; Houston: 2000. [Google Scholar]
- 22.van Dongen JT, Licausi F. Low-Oxygen Stress in Plants: Oxygen Sensing and Adaptive Responses to Hypoxia. Springer; Vienna: 2014. [Google Scholar]
- 23.Gfeller RP, Gibbs M. Fermentative metabolism of Chlamydomonas reinhardtii: I. Analysis of fermentative products from starch in dark and light. Plant Physiol. 1984;75:212–218. doi: 10.1104/pp.75.1.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Catalanotti C, Yang W, Posewitz MC, Grossman AR. Fermentation metabolism and its evolution in algae. Front Plant Sci. 2013;4:150. doi: 10.3389/fpls.2013.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Banti V, et al. Low oxygen response mechanisms in green organisms. Int J Mol Sci. 2013;14:4734–4761. doi: 10.3390/ijms14034734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blanc G, et al. The genome of the polar eukaryotic microalga Coccomyxa subellipsoidea reveals traits of cold adaptation. Genome Biol. 2012;13:R39. doi: 10.1186/gb-2012-13-5-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mus F, Dubini A, Seibert M, Posewitz MC, Grossman AR. Anaerobic acclimation in Chlamydomonas reinhardtii: Anoxic gene expression, hydrogenase induction, and metabolic pathways. J Biol Chem. 2007;282:25475–25486. doi: 10.1074/jbc.M701415200. [DOI] [PubMed] [Google Scholar]
- 28.Lou HQ, et al. A formate dehydrogenase confers tolerance to aluminum and low pH. Plant Physiol. 2016;171:294–305. doi: 10.1104/pp.16.01105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fuentes JL, et al. Phylogenetic characterization and morphological and physiological aspects of a novel acidotolerant and halotolerant microalga Coccomyxa onubensis sp. nov. (Chlorophyta, Trebouxiophyceae) J Appl Phycol. 2016;28:3269–3279. [Google Scholar]
- 30.Koechler S, et al. Arsenite response in Coccomyxa sp. Carn explored by transcriptomic and non-targeted metabolomic approaches. Environ Microbiol. 2016;18:1289–1300. doi: 10.1111/1462-2920.13227. [DOI] [PubMed] [Google Scholar]
- 31.Kunze M, Pracharoenwattana I, Smith SM, Hartig A. A central role for the peroxisomal membrane in glyoxylate cycle function. Biochim Biophys Acta. 2006;1763:1441–1452. doi: 10.1016/j.bbamcr.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 32.Ellington WR. Evolution and physiological roles of phosphagen systems. Annu Rev Physiol. 2001;63:289–325. doi: 10.1146/annurev.physiol.63.1.289. [DOI] [PubMed] [Google Scholar]
- 33.Uda K, Hoshijima M, Suzuki T. A novel taurocyamine kinase found in the protist Phytophthora infestans. Comp Biochem Physiol B Biochem Mol Biol. 2013;165:42–48. doi: 10.1016/j.cbpb.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 34.Canonaco F, Schlattner U, Pruett PS, Wallimann T, Sauer U. Functional expression of phosphagen kinase systems confers resistance to transient stresses in Saccharomyces cerevisiae by buffering the ATP pool. J Biol Chem. 2002;277:31303–31309. doi: 10.1074/jbc.M204052200. [DOI] [PubMed] [Google Scholar]
- 35.Canonaco F, Schlattner U, Wallimann T, Sauer U. Functional expression of arginine kinase improves recovery from pH stress of Escherichia coli. Biotechnol Lett. 2003;25:1013–1017. doi: 10.1023/a:1024172518062. [DOI] [PubMed] [Google Scholar]
- 36.Cullen WR, Reimer KJ. Arsenic speciation in the environment. Chem Rev. 1989;89:713–764. [Google Scholar]
- 37.Tripathi RD, et al. Arsenic hazards: Strategies for tolerance and remediation by plants. Trends Biotechnol. 2007;25:158–165. doi: 10.1016/j.tibtech.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 38.Birben E, Sahiner UM, Sackesen C, Erzurum S, Kalayci O. Oxidative stress and antioxidant defense. World Allergy Organ J. 2012;5:9–19. doi: 10.1097/WOX.0b013e3182439613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mukhopadhyay R, Rosen BP. Arsenate reductases in prokaryotes and eukaryotes. Environ Health Perspect. 2002;110:745–748. doi: 10.1289/ehp.02110s5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dhankher OP, et al. Engineering tolerance and hyperaccumulation of arsenic in plants by combining arsenate reductase and gamma-glutamylcysteine synthetase expression. Nat Biotechnol. 2002;20:1140–1145. doi: 10.1038/nbt747. [DOI] [PubMed] [Google Scholar]
- 41.Ali W, et al. Heterologous expression of the yeast arsenite efflux system ACR3 improves Arabidopsis thaliana tolerance to arsenic stress. New Phytol. 2012;194:716–723. doi: 10.1111/j.1469-8137.2012.04092.x. [DOI] [PubMed] [Google Scholar]
- 42.Kondrashov FA. Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc Biol Sci. 2012;279:5048–5057. doi: 10.1098/rspb.2012.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Šmarda P, et al. Ecological and evolutionary significance of genomic GC content diversity in monocots. Proc Natl Acad Sci USA. 2014;111:E4096–E4102. doi: 10.1073/pnas.1321152111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Milledge JJ. Microalgae–Commercial potential for fuel, food and feed. Food Sci Technol (Campinas) 2012;26:26–28. [Google Scholar]
- 45.Varshney P, Mikulic P, Vonshak A, Beardall J, Wangikar PP. Extremophilic micro-algae and their potential contribution in biotechnology. Bioresour Technol. 2015;184:363–372. doi: 10.1016/j.biortech.2014.11.040. [DOI] [PubMed] [Google Scholar]
- 46.Nishikawa K, Yamakoshi Y, Uemura I, Tominaga N. Ultrastructural changes in Chlamydomonas acidophila (Chlorophyta) induced by heavy metals and polyphosphate metabolism. FEMS Microbiol Ecol. 2003;44:253–259. doi: 10.1016/S0168-6496(03)00049-7. [DOI] [PubMed] [Google Scholar]
- 47.Minoda A, et al. Recovery of rare earth elements from the sulfothermophilic red alga Galdieria sulphuraria using aqueous acid. Appl Microbiol Biotechnol. 2015;99:1513–1519. doi: 10.1007/s00253-014-6070-3. [DOI] [PubMed] [Google Scholar]
- 48.Hirose Y, et al. Complete genome sequence of cyanobacterium Geminocystis sp. strain NIES-3708, which performs type II complementary chromatic acclimation. Genome Announc. 2015;3:e00357–e15. doi: 10.1128/genomeA.00357-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohtsubo Y, Maruyama F, Mitsui H, Nagata Y, Tsuda M. Complete genome sequence of Acidovorax sp. strain KKS102, a polychlorinated-biphenyl degrader. J Bacteriol. 2012;194:6970–6971. doi: 10.1128/JB.01848-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marçais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27:578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 52.Kosugi S, Hirakawa H, Tabata S. GMcloser: Closing gaps in assemblies accurately with a likelihood-based selection of contig or long-read alignments. Bioinformatics. 2015;31:3733–3741. doi: 10.1093/bioinformatics/btv465. [DOI] [PubMed] [Google Scholar]
- 53.Otto TD, Sanders M, Berriman M, Newbold C. Iterative correction of reference nucleotides (iCORN) using second generation sequencing technology. Bioinformatics. 2010;26:1704–1707. doi: 10.1093/bioinformatics/btq269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li H, et al. 1000 Genome Project Data Processing Subgroup The sequence alignment/map (SAM) format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stanke M, Diekhans M, Baertsch R, Haussler D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008;24:637–644. doi: 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- 59.Kent WJ. BLAT–The BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.R Core Team 2013. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna), Version 2.14.2.
- 63.Zheng W, Chung LM, Zhao H. Bias detection and correction in RNA-sequencing data. BMC Bioinformatics. 2011;12:290. doi: 10.1186/1471-2105-12-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Risso D, Schwartz K, Sherlock G, Dudoit S. GC-content normalization for RNA-seq data. BMC Bioinformatics. 2011;12:480. doi: 10.1186/1471-2105-12-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 66.Lauersen KJ, et al. Peroxisomal microbodies are at the crossroads of acetate assimilation in the green microalga Chlamydomonas reinhardtii. Algal Res. 2016;16:266–274. [Google Scholar]
- 67.Posewitz MC, et al. Discovery of two novel radical S-adenosylmethionine proteins required for the assembly of an active [Fe] hydrogenase. J Biol Chem. 2004;279:25711–25720. doi: 10.1074/jbc.M403206200. [DOI] [PubMed] [Google Scholar]
- 68.Hirooka S, Higuchi S, Uzuka A, Nozaki H, Miyagishima SY. Acidophilic green alga Pseudochlorella sp. YKT1 accumulates high amount of lipid droplets under a nitrogen-depleted condition at a low-pH. PLoS One. 2014;9:e107702. doi: 10.1371/journal.pone.0107702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Minoda A, Sakagami R, Yagisawa F, Kuroiwa T, Tanaka K. Improvement of culture conditions and evidence for nuclear transformation by homologous recombination in a red alga, Cyanidioschyzon merolae 10D. Plant Cell Physiol. 2004;45:667–671. doi: 10.1093/pcp/pch087. [DOI] [PubMed] [Google Scholar]
- 70.Nakazawa A, Nozaki H. Phylogenetic analysis of the tetrasporalean genus Asterococcus (Chlorophyceae) based on 18S ribosomal RNA gene sequences. Shokubutsu Kenkyu Zasshi. 2004;79:255–261. [Google Scholar]
- 71.Nollet LML, De Gelder LSP. Handbook of Water Analysis. 3rd Ed Taylor & Francis Group; Boca Raton, FL: 2014. [Google Scholar]
- 72.Murota C, et al. Arsenic tolerance in a Chlamydomonas photosynthetic mutant is due to reduced arsenic uptake even in light conditions. Planta. 2012;236:1395–1403. doi: 10.1007/s00425-012-1689-8. [DOI] [PubMed] [Google Scholar]
- 73.Yoon OK, Brem RB. Noncanonical transcript forms in yeast and their regulation during environmental stress. RNA. 2010;16:1256–1267. doi: 10.1261/rna.2038810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fujiwara T, et al. A nitrogen source-dependent inducible and repressible gene expression system in the red alga Cyanidioschyzon merolae. Front Plant Sci. 2015;6:657. doi: 10.3389/fpls.2015.00657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. [Google Scholar]
- 76.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 78.Tanabe AS. Kakusan4 and Aminosan: Two programs for comparing nonpartitioned, proportional and separate models for combined molecular phylogenetic analyses of multilocus sequence data. Mol Ecol Resour. 2011;11:914–921. doi: 10.1111/j.1755-0998.2011.03021.x. [DOI] [PubMed] [Google Scholar]
- 79.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 80.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 81.Durzan DJ. Automated chromatographic analysis of free monosubstituted guanidines in physiological fluids. Can J Biochem. 1969;47:657–664. doi: 10.1139/o69-101. [DOI] [PubMed] [Google Scholar]
- 82.Bennett MS, Guan Z, Laurberg M, Su XD. Bacillus subtilis arsenate reductase is structurally and functionally similar to low molecular weight protein tyrosine phosphatases. Proc Natl Acad Sci USA. 2001;98:13577–13582. doi: 10.1073/pnas.241397198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The C. eustigma NIES-2499 whole-genome and gene models have been deposited in DNA Data Bank of Japan (DDBJ)/European Molecular Biology Laboratory (EMBL)/GenBank under the accession code PRJDB5468. The dataset includes sequences of the nuclear and mitochondrial genomes. Because the chloroplast genome was highly repetitive and the genome could not be assembled well, the chloroplast genome was omitted from the dataset. The RNA-seq data of C. eustigma and C. reinhardtii have been deposited in DDBJ/EMBL/GenBank (accession codes PRJDB6154 and PRJDB6155, respectively).