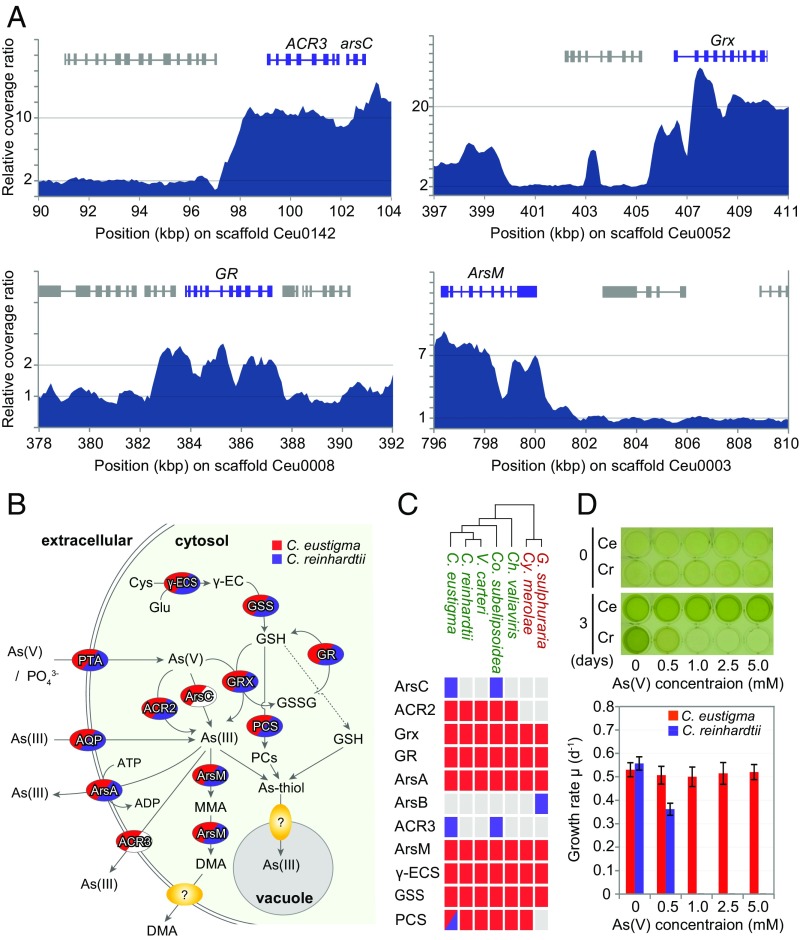
Fig. 6.
Acquisition through HGT and expansion of genes in the arsenic biotransformation and detoxification system in C. eustigma. (A) Relative sequencing coverage ratio of Illumina MiSeq reads (200-bp window with 100-bp overlap) showing the copy numbers of arsenic biotransformation and detoxification genes. (B) Overview of the arsenic biotransformation and detoxification system (37). As(V) and As(III) are taken up through phosphate transporter (PTA) as a phosphate analog and through aquaglycoporins (AQP), respectively. As(V) is reduced to As(III) by glutaredoxin (Grx) by using glutathione (GSH) as a reductant or arsenate reductase ArsC or ACR2. As(III) is excreted by As(III)-specific transporter ArsA or ACR3. GSH is an antioxidant and is synthesized by γ-glutamylcysteine synthetase (γ-ECS) and glutathione synthetase (GSS). The methylation of As(III) to monomethylarsonic acid (MMA) and dimethylarsinic acid (DMA) is also thought to function as inorganic arsenic biotransformation and detoxification. As(III) is chelated by phytochelatins (PCs), which are synthesized by phytochelatin synthase (PCS) or GSH to form the As(III)-thiol complex, which is finally transported into the vacuole. (C) Presence or absence of arsenic biotransformation and detoxification genes in the genomes of five green algae (shown in green) and two thermo-acidophilic red algae (shown in red). The red and blue boxes indicate the presence of the gene; blue boxes indicate a gene that was horizontally transferred. The gray boxes indicate the absence of the gene. (D) Effects of the concentration of arsenate As(V) on the growth of C. eustigma and C. reinhardtii. Cells were cultured in modified original medium containing both 1 mM phosphate (originally 10 mM) and various concentrations of arsenate for 3 d. The error bars in the graph represent the SD of three biological replicates.