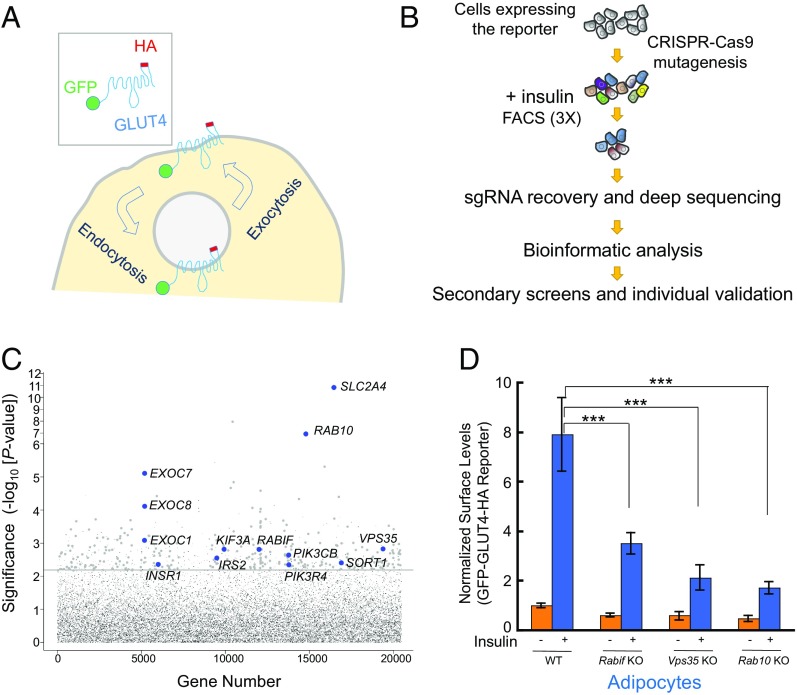
Fig. 1.
Dissection of insulin-stimulated GLUT4 exocytosis using CRISPR genetic screens. (A) Diagram of the GFP-GLUT4-HA reporter used to monitor insulin-dependent GLUT4 trafficking. After the translocation of the reporter, the HA epitope is exposed to the cell exterior (60). (B) Illustration of the genome-wide genetic screen of insulin-stimulated GLUT4 exocytosis. (C) Ranking of genes in the CRISPR screen based on the P value. Each dot represents a gene. Genes above the horizontal line were tested in the pooled secondary screen. A gene is shown as a large dot if it was validated in the secondary screen. Other genes are shown as small gray dots. Selected known or validated regulators of GLUT4 exocytosis are labeled. (D) Selected genes from the screen were individually mutated in mouse adipocytes using CRISPR-Cas9. Effects of the mutations on insulin-stimulated GLUT4 exocytosis were measured by flow cytometry. Error bars indicate SD. ***P < 0.001.