Figure 1. Design and testing of asymmetric nucleosomes.
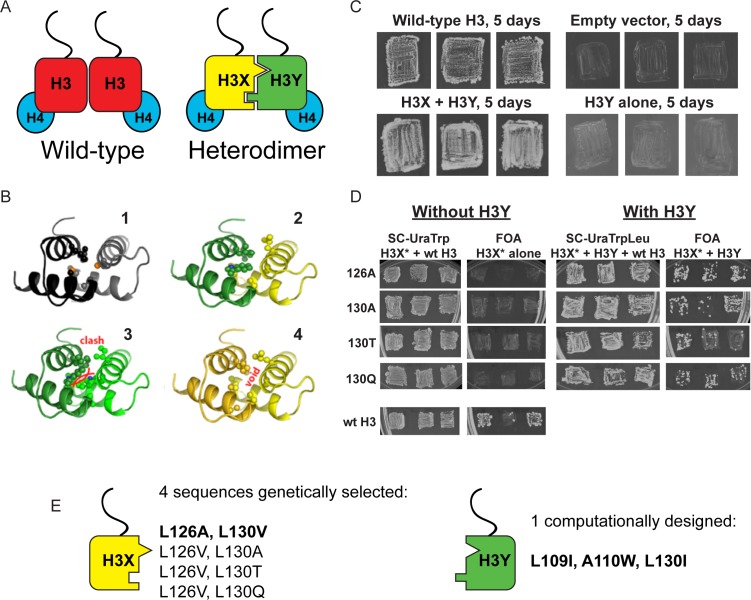
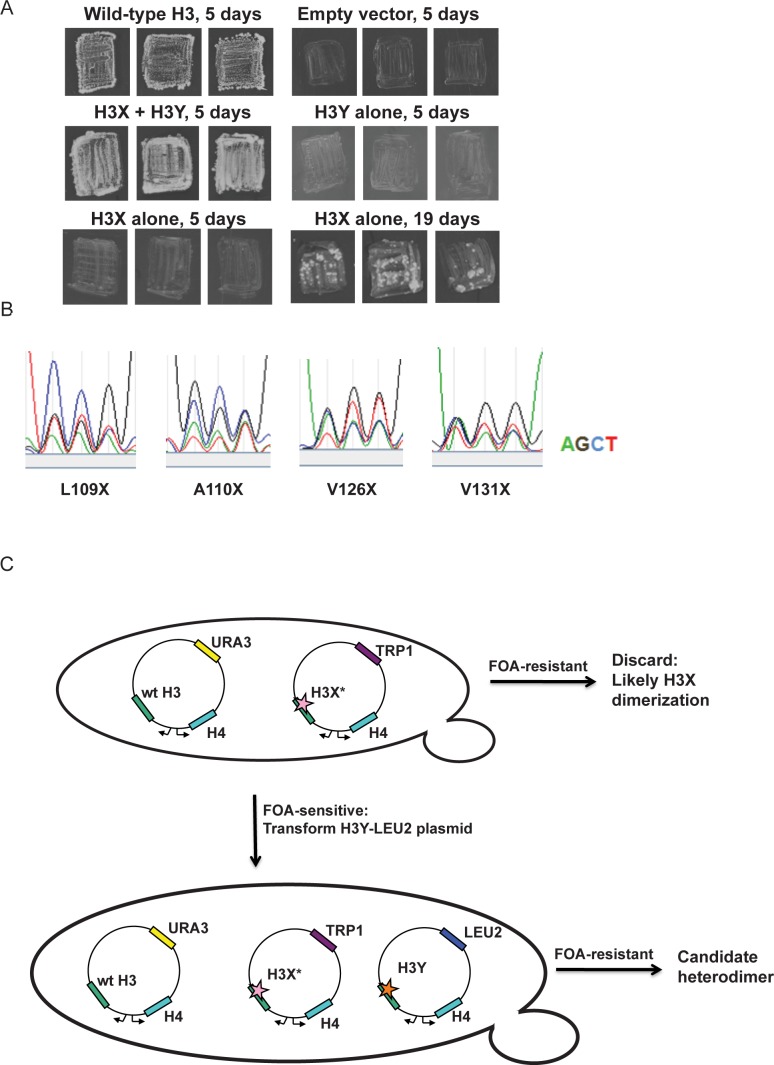
(A) Schematic of asymmetric histone H3 design. Left: Wild-type H3/H4 tetramers are symmetrical, with H3-H3 interactions serving as the dimerization interface. The H3 N-terminus, site of many modifiable residues, is indicated protruding from the globular domain. Right: Obligate heterodimer H3s are comprised of distinct ‘X’ and ‘Y’ interaction partners which are altered to prevent either H3 from homodimerizing. (B) Computational models of the designed histone H3 heterodimer. (1) The four-helix bundle comprising the H3-H3 C-terminal dimerization interface (pdb 1KX3). Leucine and alanine residues at the hydrophobic interface are indicated. (2) The designed asymmetric mutations pack efficiently as a heterodimer, with partners colored in green (H3Y) and yellow (H3X). Indicated residues at the H3-H3 interface were engineered to form a bump-hole interface to increase interaction affinity. (3-4) Altered H3s are designed not to homodimerize due to van der Waals clashes (3) or voids (4) in the hydrophobic core. (C–D) Genetic analysis of heterodimeric H3X/H3Y pairs. C)H3Y alone cannot support growth. Images show growth of yeast carrying wild-type H3 on a URA3-marked plasmid as well as either empty plasmid, wild-type H3, H3Y alone, or both H3X and H3Y. Three independent transformants for each strain were grown on 5-FOA to select against the URA3 shuffle plasmid. (D) H3X alone cannot support growth. Left panels: growth of three independent transformants of various H3X strains (evolved second-generation mutations indicated at left) in the absence of H3Y. Bottom: positive control with a TRP1-marked wild-type histone plasmid. Right panels: growth of indicated H3X strains in the presence of H3Y. Synthetic complete (SC) plates were grown 3 days, FOA plates 9 days. No growth on FOA was observed for any of the four evolved H3X alleles in the absence of H3Y, even after extended incubation. (E) Sequences of final H3X-H3Y molecules. All four H3X variants have been validated genetically, while two variants – 126A/130V and 126V/130A – have been validated biochemically (see Figure 2 and not shown). 126A is used for all functional analyses.