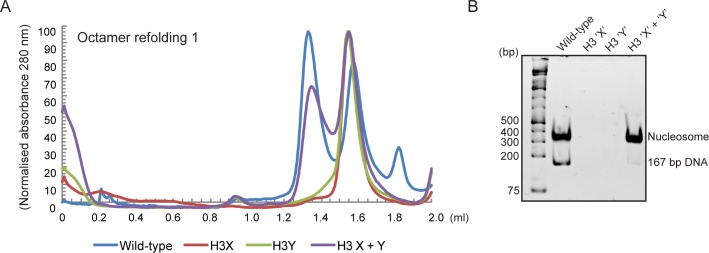
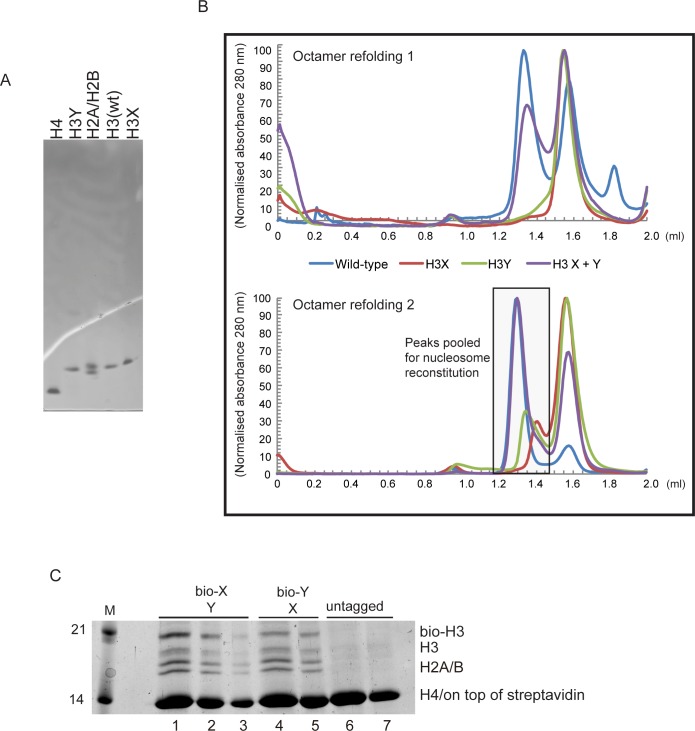
Figure 3. Human histone H3 mutants ‘X’ and ‘Y’ form obligate heterodimers in vitro.
(A) Size exclusion (Superdex 200 Increase 3.2/300) gel filtration profiles showing the purification of octamer refolding reactions containing wild-type and heterodimer ‘X’ and ‘Y’ histone H3. Octamer refolding in the presence of both ‘X’ and ‘Y’ H3 yields histone octamers with a characteristic elution profile, similar to refolding reactions containing wild-type H3, while reconstitutions with only H3X or only H3Y form aggregates. (B) Native PAGE analysis of nucleosome reconstitution experiments with purified histones from (A). H3X/Y octamer reconstitutions readily form nucleosomes when assembled onto DNA, while the aggregates formed in octamer refolding reactions containing either ‘X’ or ‘Y’ alone do not form histone octamers and therefore cannot form nucleosomes in vitro.