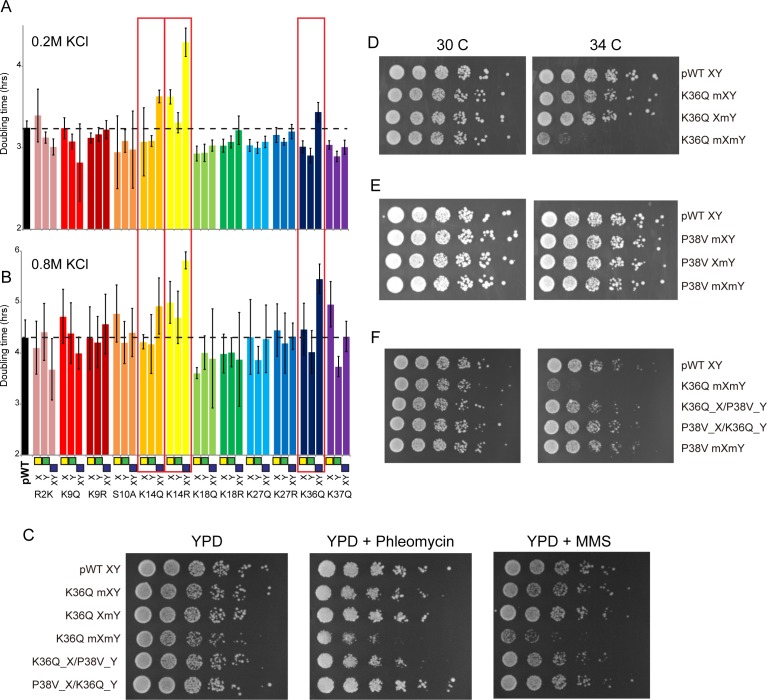
Figure 4. Genetic epistasis between the H3 tails.
(A–B) We constructed 12 sets of point mutations in histone H3, with each mutant cycle including H3Xmut/H3Ywt, H3Xwt/H3Ymut, and H3Xmut/H3Ymut – for each mutant trio, X/Y/double mutations are ordered from left to right, as indicated with green/yellow/blue boxes in Panel (B). For all 12 trios, as well as the matched ‘pseudo-wt’ (labeled as pWT) carrying H3Xwt/H3Ywt (leftmost bar and dashed line), we measured doubling times in at least six replicate cultures in low (A) and high (B) KCl. Boxes emphasize H3K14R and H3K36Q trios, as detailed in text. Note that growth rates here were measured robotically in a 96-well format, so doubling times are slower than those typically measured in well-aerated cultures (Figure 4—figure supplement 1A). (C) Effects of H3K36Q mutants on DNA damage sensitivity. Serial dilutions of the indicated strains were plated on YPD, YPD + 10 ng/ml phleomycin or YPD + 0.025% MMS, as indicated. pWT XY (PKY4704), K36Q mXY (PKY4829), K36Q XmY (PKY4831), K36Q mXmY (PKY4834), K36Q_X/P38V_Y (PKY5138), and P38V_X/K36Q_Y (PKY5140). (D–F) Serial dilution growth assay for the indicated strains incubated at 30 C or 34 C, as indicated. Note that yeast carrying pseudo-wild type H3X/H3Y nucleosomes grow poorly at 37C, so we assayed temperature sensitivity at 34 C. Mutant set in (D) shows temperature-sensitivity is specific to symmetric H3K36Q mutant. Mutant set in (E) shows that complete loss of H3K36me3 in the symmetric H3P38V mutant (where H3K36me3 is replaced with H3K36me2) is compatible with rapid growth at high temperatures. Mutant set in (F) shows that a single dimethylated K36 residue per nucleosome is compatible with rapid growth at high temperatures. Strains analyzed were: pWT XY (PKY4704), R2K mXY (PKY4749), R2K XmY (PKY4751), R2K mXmY (PKY4753), K9Q mXY (PKY4714), K9Q XmY (PKY4715), K9Q mXmY (PKY4706), K9R mXY (PKY4773), K9R XmY (PKY4775), K9R mXmY (PKY4777), S10A mXY (PKY4716), S10A XmY (PKY4717), S10A mXmY (PKY4707), K14Q mXY (PKY4789), K14Q XmY (PKY4791), K14Q mXmY (PKY4793), K14R mXY (PKY4781), K14R XmY (PKY4783), K14R mXmY (PKY4786), K18Q mXY (PKY4805), K18Q XmY (PKY4807), K18Q mXmY (PKY4809), K18R mXY (PKY4797), K18R XmY (PKY4799), K18R mXmY (PKY4801), K27Q mXY (PKY4822), K27Q XmY (PKY4823), K27Q mXmY (PKY4825), K27R mXY (PKY4813), K27R XmY (PKY4815), K27R mXmY (PKY4817), K36Q mXY (PKY4829), K36Q XmY (PKY4831), K36Q mXmY (PKY4834), K37Q mXY (PKY4837), K37Q XmY (PKY4839), K37Q mXmY (PKY4841), P38V mXY (PKY5033), P38V XmY (PKY5035), P38V mXmY (PKY5037), K36Q mX/P38V mY (PKY5138), and P38V mX/K36Q mY (PKY5140). pWT XY, pseudo WT; mXY, mutation on X; XmY, mutation on Y; and mXmY, mutation on both X and Y.
Figure 4—figure supplement 1. Significant growth defects for the H3K36Q double mutant, relative to either single mutant.
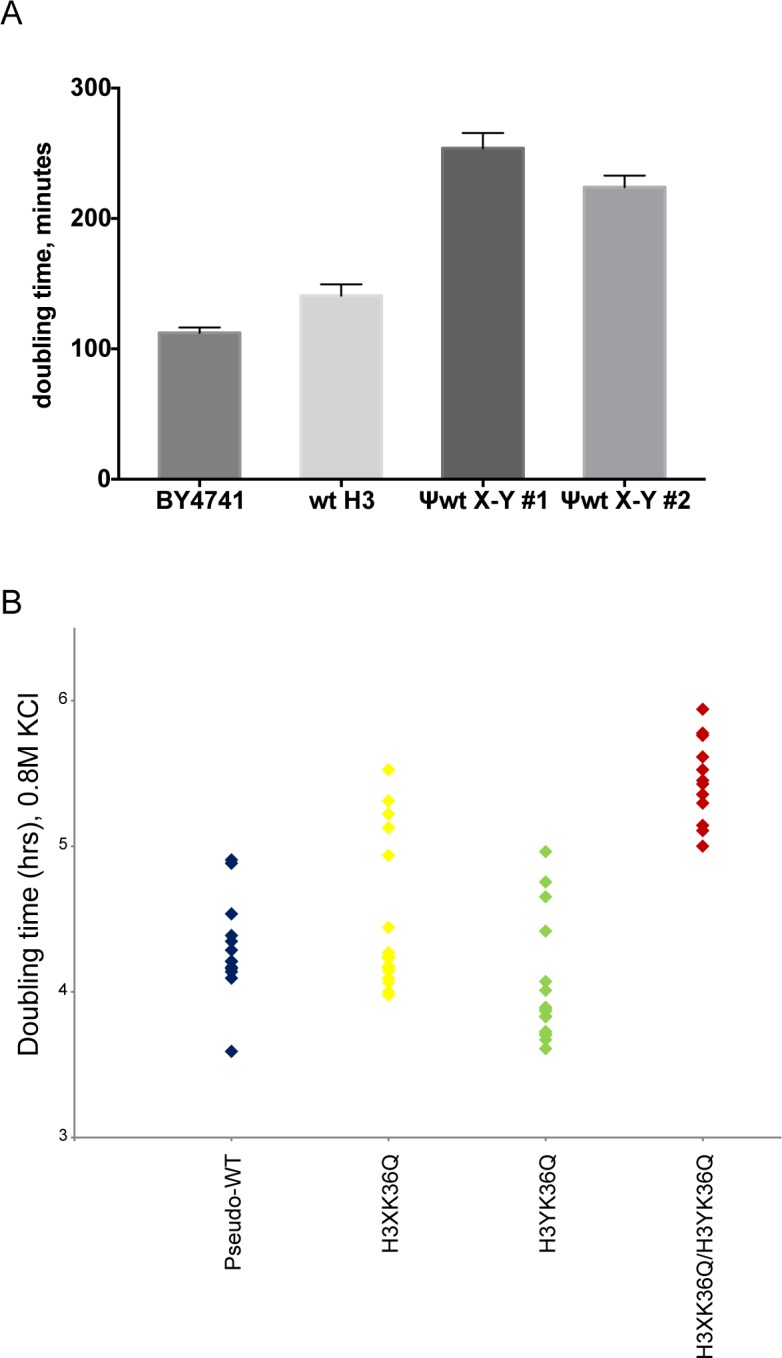
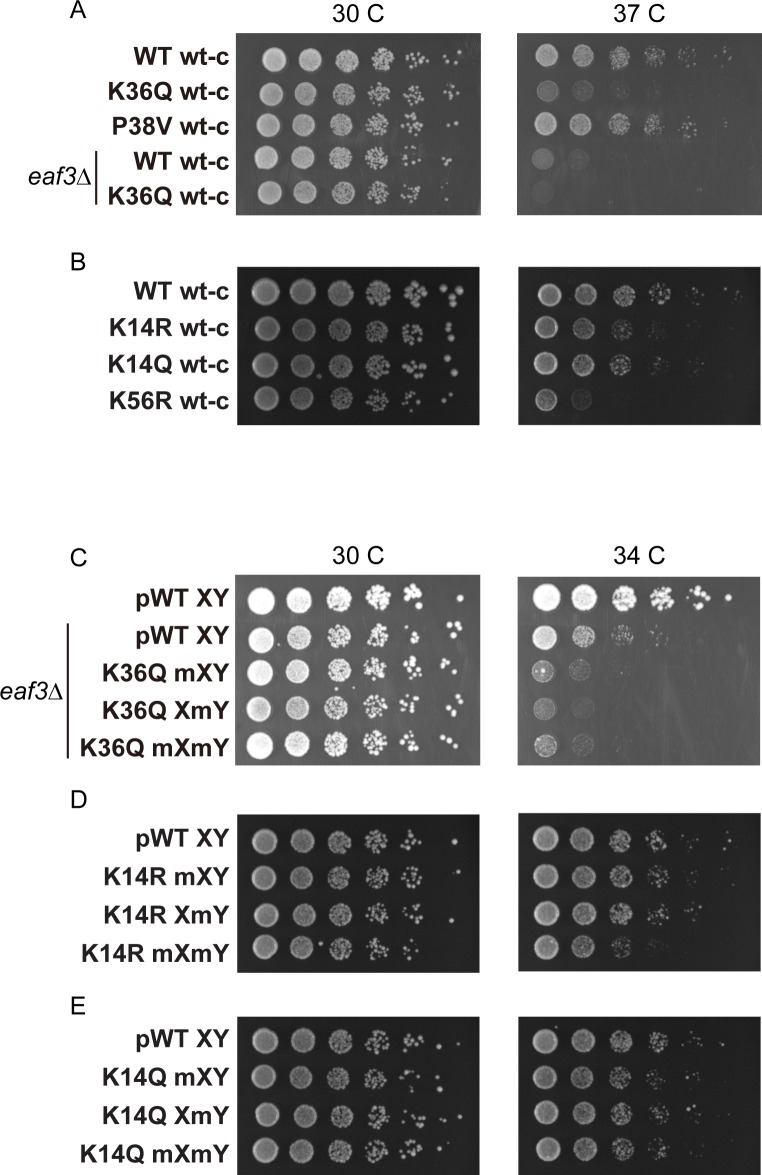
Figure 4—figure supplement 2. Temperature sensitivity of various histone mutant strains.
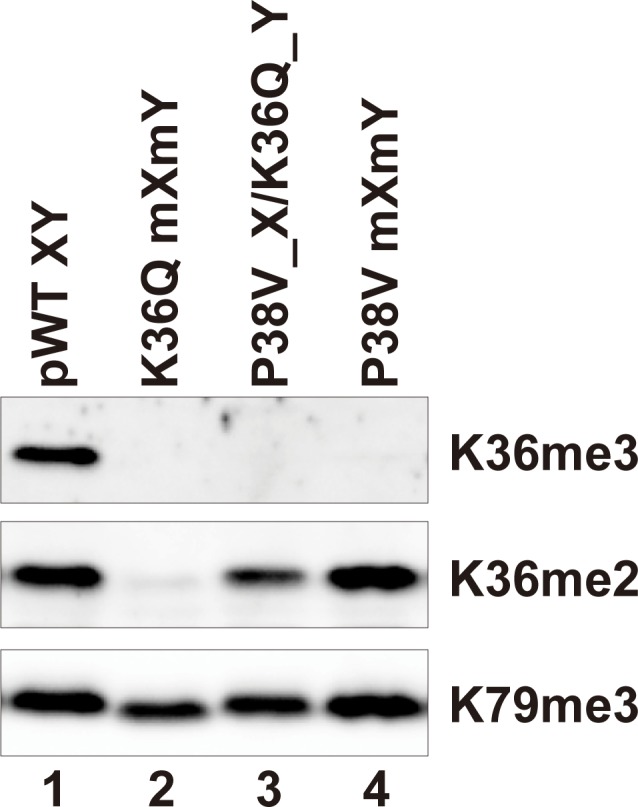
Figure 4—figure supplement 3. H3P38V mutations block trimethylation but not dimethylation of H3K36.