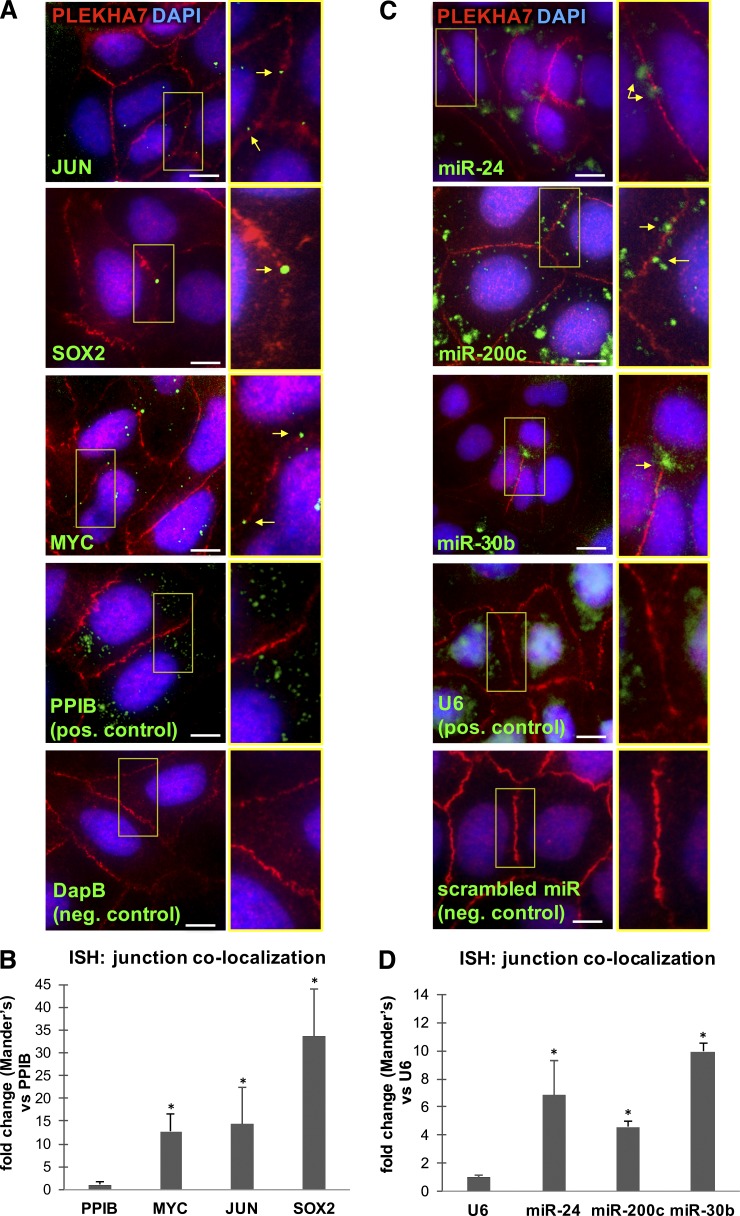
Figure 3.
PLEKHA7-associated RNAs localize at the ZA. (A) ISH of Caco2 cells for MYC, JUN, and SOX2 mRNAs; an ER-specific PPIB probe was used as the positive control and a bacterial DapB RNA probe as the negative control. (B) Quantification of mRNA colocalization to the junctions by using the Manders coefficient, expressed as fold difference of the junctional colocalization of the PPIB control (mean ± SEM from n = 6 fields; *, P < 0.01, Student’s two-tailed t test). (C) ISH of Caco2 cells for miR-24, miR-200c, and miR-30b miRNAs; a nuclear-specific U6 small RNA probe was used as the positive control and a scrambled miRNA probe as the negative control. (D) Quantification of miRNA colocalization to the junctions by using the Manders coefficient, expressed as fold difference of the junctional colocalization of the U6 control (mean ± SEM from n = 6 fields; *, P < 0.04, Student’s two-tailed t test). In all cases, ISH was followed by PLEKHA7 co-immunofluorescence and DAPI staining. Merged images are shown. Enlarged areas indicated by yellow boxes are shown to the right of each image. Arrows indicate hybridization signals. Bars, 20 µM. Insets are magnified 2×.