Figure 7. PR diminishes recruitment of STAT2/IRF9 to ISG promoters.
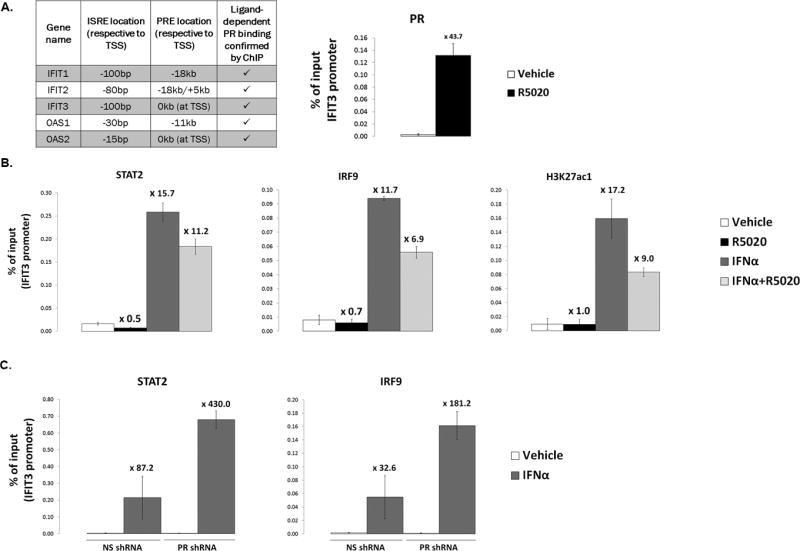
(A). Left: table summary of ISG promoters/enhancers assayed by ChIP-qPCR for PR, STAT2, and IRF9 recruitment. Location of interferon-stimulated response elements (ISREs) and progesterone receptor response elements (PREs) from the transcriptional start site (TSS) of each gene is shown in kilobases (kb). Right: T47D-co cells were serum-starved for 18hr. Cells were then treated with 10nM R5020 or vehicle for 30min. Fixed lysates were subjected to ChIP with antibodies against PR or a species-specific IgG (control; not shown), and qPCR was performed on the isolated DNA using primers designed to amplify the IFIT3 promoter. A percentage of ChIP’d DNA over input DNA is shown. (B). T47D-co cells were serum-starved for 18hr. Cells were then treated with 10nM R5020, 1000IU/ml IFNα, or a co-treatment of both (or appropriate vehicle controls). Fixed lysates were subjected to ChIP with antibodies against STAT2, IRF9, H3K27ac1 or a species-specific IgG (control; not shown), and qPCR was performed on the isolated DNA using primers designed to amplify the IFIT3 promoter. A percentage of ChIP’d DNA over input DNA is shown. (C). ChIP experiments were performed as in (B), but T47D-co NS and PR shRNA cells were used, with only IFNα treatment. All ChIP experiments were performed in triplicate; a representative experiment is shown here. Fold-recruitment in treated conditions (R5020, IFNα, or combo), as compared to vehicle treatment, is displayed above each bar. Error bars represent SD of technical replicates.
