Figure 1.
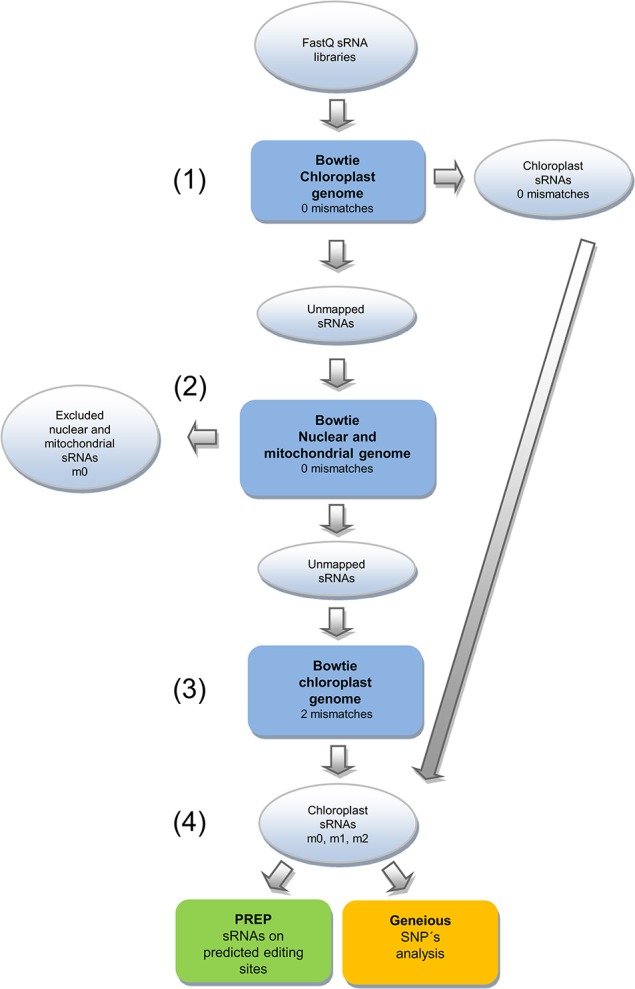
Pipeline for identification of editing sites using chloroplast RNA transcripts. (1) sRNA-seq/mRNA-seq reads were filtered by mapping against the chloroplast reference genome. Mapped reads were saved as another file named as m0 (chloroplast RNAs m0). (2) Reads that did not map were subjected to a new round of mapping against nuclear and mitochondrial reference genomes, and those reads that did map were discarded. (3) The remaining unmapped reads were remapped against the chloroplast genome allowing up to 2 mismatches using Bowtie. (4) The resulting mapped reads (chloroplast m0 + m2), plus the m0 file, were used in the analysis to predict transcript editing sites through PREP and Geneious SNPs approaches.
