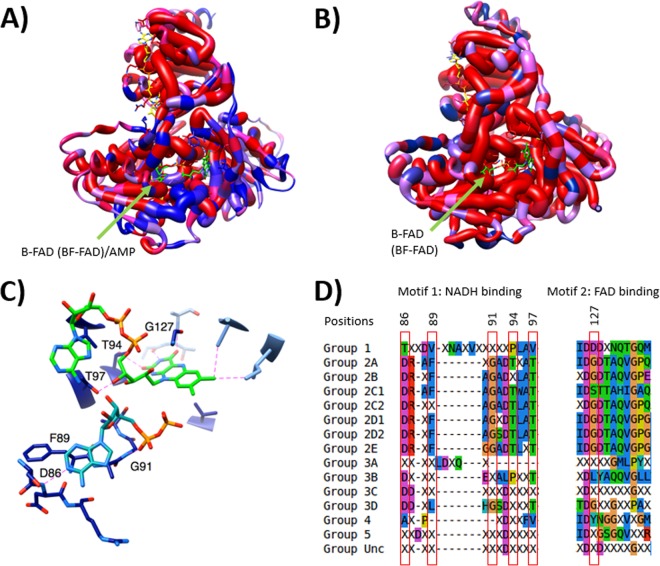
FIG 5.
Structural analyses of Bf and non-Bf Etfs showing differences near the β-FAD site. (A) Homology model of Etf-α and Etf-β with mapped conserved differences in amino acid type between G1 Etfs, with representatives that do not bifurcate and contain AMP in the β subunit, and G2 Etfs, with representatives that have been demonstrated to catalyze electron-bifurcating reactions. Red indicates equivalent residues at a position, whereas blue indicates a position at which the distribution of residues characterizing one group differs from the distribution of residues found in the other group. The thickness of the worms indicates whether amino acids at a position share the same functional property (wide worm) or not (thin worm) within G2. (B) Homology model of Etf-α/β with mapped conserved differences in amino acid use between Etfs in G2A and G2B combined, which have representatives known to bifurcate, and Etfs in G2D2, which includes the FixAB Etfs. Note the high degree of conservation (thick red worms) around the putative β-FAD binding site, leading us to predict that G2D2 Etfs also bifurcate. Thick worms indicate high levels of conservation within G2D2. (C) Closeup depiction of the NADH-binding (blue green) and FAD-binding (bright green) residues in bifurcating Etfs. Key residues proposed to coordinate these two molecules are numbered with their positions in the sequence of R. palustris Etf (GenBank accession no. YP_783418). Only a portion of the NADH molecule, where the quality of the electron density was good enough to permit unambiguous refinement in the structure upon which our model is based (PDB accession no. 4L2I), is shown. Only residues associated with the proposed ability to bifurcate are shown. (D) Alignment of residues predicted to coordinate NADH and FAD in bifurcating Etfs. Groups in the first column correspond to the groups mentioned in Table 1. Unc indicates the uncharacterized group. Residues highlighted in panel C are shown in red boxes.