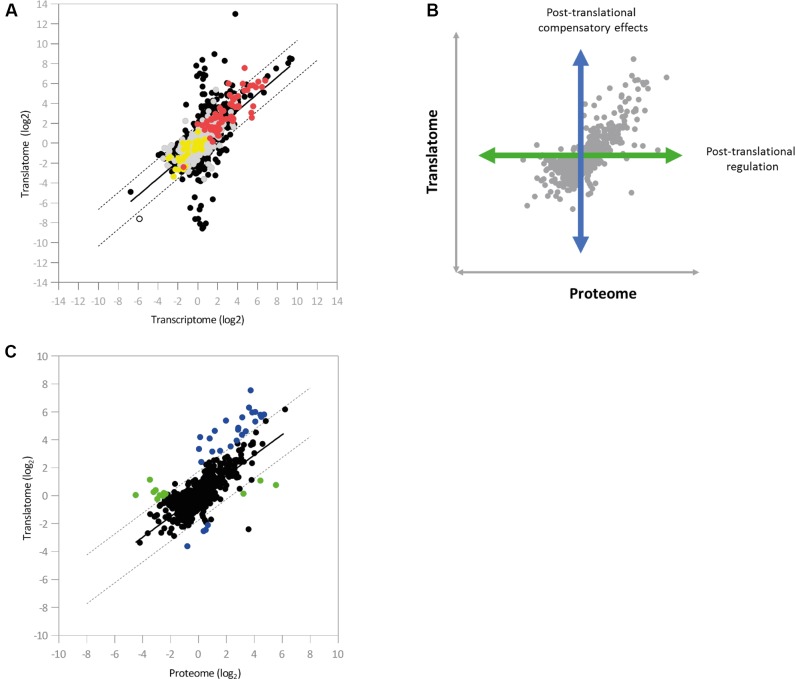
FIGURE 4.
Integration of the regulatory datasets. (A) Color-coded integration of proteomic data into the pairwise comparison between the Hfq transcriptome and translatome (n = 5910). Red and yellow dots indicate loci that show up- and down-regulated protein abundance, respectively, in the Δhfq mutant. Loci exhibiting no significant change in protein abundance are indicated in gray (Supplementary Table S4). (B) Illustration of the effects of post-translational control on protein abundance. (C) Scatter-plot showing the pairwise comparisons of log2 ratios between the Δhfq translatome and proteome (n = 1867). Post-translationally regulated loci are marked in green, while loci displaying compensatory post-translational effects are marked in blue (Supplementary Table S4). In both cases, dashed lines indicate two-fold ratios of differential expression from the regression line (in black).