Figure 2.
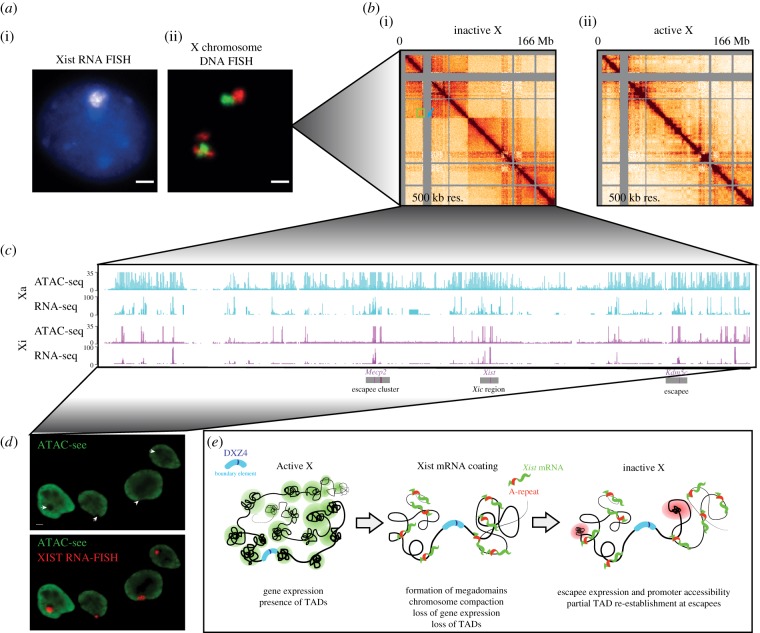
Zooming in on chromosome structure on the Xi. (a) (i) Xist RNA FISH in female mouse NPCs showing a cloud of Xist RNA coating the X-chromosome. (ii) DNA FISH on the X-chromosome showing the separation of the Xi into two domains or lobes. The Xa, shown for comparison, shows mixing of the two domains. (b) Allele-specific HiC at 500 kb resolution for the Xi (i) and Xa (ii) in NPCs. The Xi is configured in two megadomains within which TAD structure is lost. The megadomains are separated by the DXZ4 satellite element (blue arrow). A few mini-TADs are re-formed on the Xi (green box). For comparison, the Xa does not have this megadomain structure, but maintains autosome-like TAD structure. (c) Allele-specific ATAC-seq and RNA-seq in NPCs on the Xa (top) and Xi (bottom). There is a global reduction in accessibility and gene expression on the Xi compared to the Xa. Regions that retain acccessibility are located at the promoters and CTCF sites close to escape genes. A cluster of escapees near Mecp2, the Xic and the Kdm5c escape locus, are indicated below. (d) ATAC-see reveals the spatial organization of accessible chromatin in NPCs. Xist RNA FISH cloud falls in a region of depletion of accessible chromatin. (e) Model for how the megadomain structure of the Xi is formed during X inactivation. TADs on the Xa are lost and replaced by two large domains separated by the DXZ4 element. Full-length Xist containing the A-repeat region is required for this restructuring. After X inactivation, small escape TADs form. Within these TADs, escape genes make contact with one another and are regulated at highly proximal CTCF elements. Figures adapted from Giorgetti et al. [68] and Chen et al. [69].