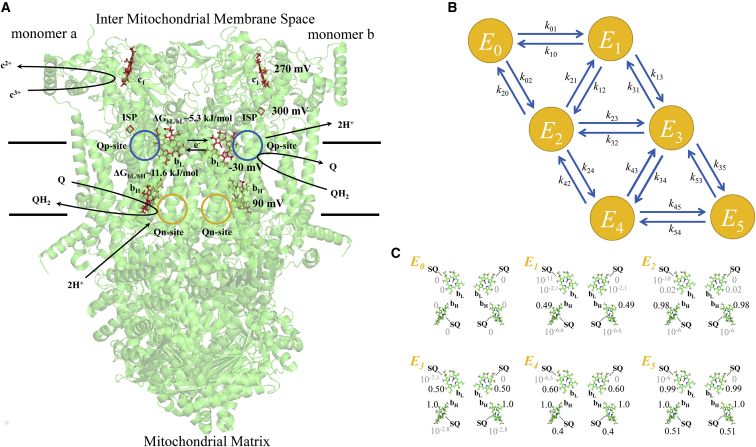
Figure 1.
Model diagram of the bc1 dimer. (A) Dimeric model of the bc1 complex is shown with major redox centers and partial reactions. Midpoint potentials for the redox centers on the complex are from Table S1 and represented at pH 7. The model assumes only one Qp site is active per turnover (31). Two turnovers at the Qp site are required per turnover at the Qn site. The model lumps oxidation of QH2 at the Qp site into a two-electron step. The first electron is used to reduce cytochrome c, and the second electron is deposited on the bc1 complex. The monomer order during QH2 oxidation at the Qp site is random. Quinone reduction at the Qn site is also random. Electrons on the complex distribute themselves among the redox centers according to the Boltzmann distribution. Electron transport between b hemes and the Qn site is electrogenic. Coulombic interaction energies between intramonomer bL and bH hemes and intermonomer bL and bL hemes are included. Blue and yellow circles are approximate locations of Qp- and Qn-site binding pockets, respectively. The dimer cartoon was generated from the crystal structure by Esser et al. (13) (PBD: 5KLV). The depicted proton uptake and release pathways are only for visual purposes. (B) State representation of the model is shown where Ei is the ith electronic state corresponding to the number of electrons residing on the complex. The state-transition rate constants, kij, are given in the Supporting Material. Probabilities <0.4 are shown in gray. (C) For each enzyme state, Ei, the probability of finding an electron on each redox center in the model is shown for the following conditions: Q pool 10% reduced, membrane potential of 0 mV, pH 7 on both sides of the membrane, no cytochrome c present, and under anoxia. Cartoon rendering of the bc1 complex was done using the software PyMOL (108). To see this figure in color, go online.