Figure 1.
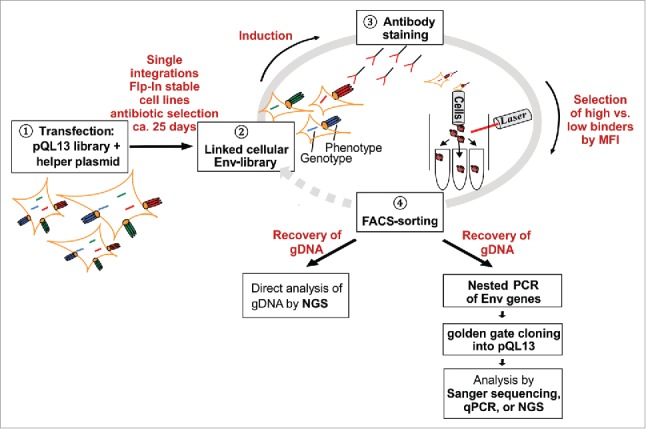
Schematic overview of the flow cytometry-based panning procedure. Stable cell lines are generated by transfection of FlpIn™ T-REx™ 293 cells with pQL13-based Env constructs and the helper plasmid pOG44 carrying the integrase (1). Although multiple Env variants can theoretically enter the human cells and be expressed for a short period of time, a linkage of geno- and phenotype is ultimately achieved because FlpInTM T-RExTM 293 cells possess precisely one FRT site, resulting in single-integration stably transfected HEK293 cells (2). After induction of Env expression, the mammalian cell library is stained with the screening antibody (3), and then envelope-expressing cells yielding the highest or lowest signal to the applied antibody are selected via fluorescence-activated cell sorting (4). For analytical purposes, genomic DNA is recovered from the selected HEK293 cells, envelope genes are amplified by nested PCR, re-cloned into pQL13 and analyzed by qPCR or sequencing of plasmid DNA recovered from single E. coli clones. In the case of larger libraries, genomic DNA from sorted HEK293 cells can be directly subjected to NGS to calculate enrichment or depletion factors for the respective envelope variants. To further enhance enrichment rates, sorted HEK293 cells could be re-expanded to be subjected to a new round of panning.
