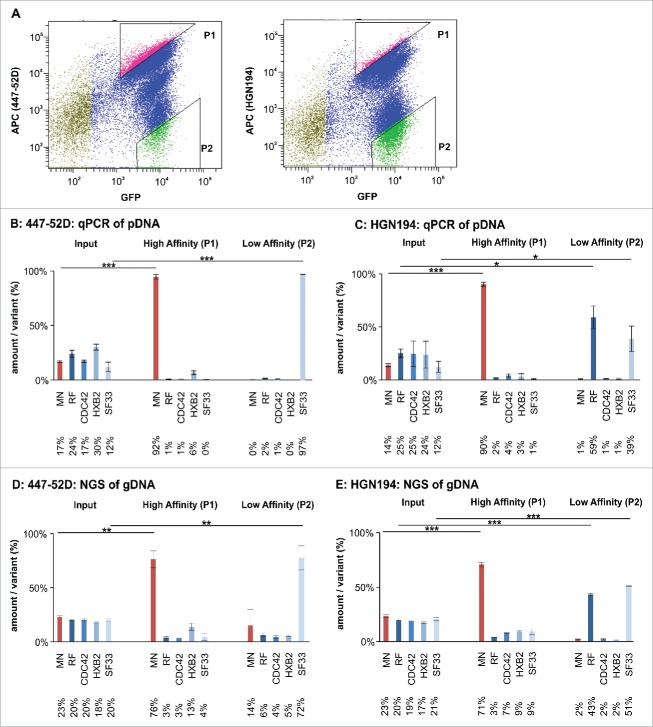
Figure 6.
Flow cytometry-based panning using the Env/V3 model library stable cell line. A: A representative sorting experiment is shown to illustrate the flow cytometry-based cell sorting strategy (50,000 events). Living, single HEK293 cells were gated according to common hierarchical gating strategies. The remaining cells were then gated for highest APC signals (447–52D or HGN194 with anti-human-APC secondary antibody) in relation to eGFP signals (expression control). Thus, a triangular gate (gate P1) was chosen to select for high affinity binders. Similarly, P2 was chosen to select for low affinity binders to 447–52D and HGN194. B to E: The stable cell line pool containing the complete 5 member Env/V3 model library was used in a flow cytometry-based panning procedure 24 h after induction with doxycycline using 2.7 nM 447–52D (B, D) or 6.7 nM HGN194 (C, E). The envelope genes from the cell line pool prior (Input) and after selection (High Affinity P1 and Low Affinity P2) were PCR-amplified from the genomic DNA, cloned into pQL13, recovered from E. coli and analyzed by qPCR (B and C) or, alternatively, the gDNA of the HEK293 cells was directly subjected to NGS analysis (D and E). The mean values of 3 independent experiments are shown. The percentages of the variants in the input mixture and after one round of panning are given. Statistics were calculated using an unpaired t-test. Asterisks are indicated for *** p < 0.001, ** p < 0.01 and * p < 0.05.