Abstract
Platinum resistance is a critical barrier for clinicians to improve the survival of ovarian cancer. Our study evaluated the correlation between copy number variations (CNVs) of neurotrophic tyrosine receptor kinase 3 (NTRK3) and the prognosis of ovarian cancer, which might predict platinum resistance in ovarian cancer patients.
Array comparative genomic hybridization (CGH) was used to test gene backgrounds between platinum-sensitive and platinum-resistant relapsed populations and CNVs of NTRK3 were indicated by cluster analysis. Fluorescence in situ hybridization (FISH) was adopted in 41 cases for further verification, which confirmed the results of array CGH. Spearman's rank correlation analysis and χ2 test were used to evaluate the accuracy of CNVs of NTRK3 which predicted platinum-sensitive or platinum-resistant recurrence.
We detected CNVs of NTRK3 between 2 groups by array CGH, and amplification of NTRK3 was confirmed by FISH in the platinum-sensitive recurrence group with enlarged samples. The test concordance of 2 methods was 78.6%. Among 41 cases with satisfied FISH results, the median time to recurrence (TTR) of patients with amplified and nonamplified NTRK3 were respectively 18 and 5 months (P <.01). The cut-off value of TTR to differentiate platinum-sensitive or platinum-resistant recurrence was 6 months in accordance with clinical practice. According to the above standard, 15 cases with NTRK3 amplification were platinum-sensitive and 12 cases without NTRK3 amplification were platinum-resistant recurrences which demonstrated that the accuracy of NTRK3 amplification/nonamplification to predict recurrent types was 65.9% (27/41).
CNVs of NTRK3 were associated with platinum-sensitive and platinum-resistant recurrences. Amplification of NTRK3 perfectly predicted platinum-sensitive relapse of ovarian cancer.
Keywords: copy number variations, neurotrophic tyrosine receptor kinase 3, platinum-resistant recurrence, platinum-sensitive recurrence
1. Introduction
Ovarian cancer is one of the most deadly gynecological cancers with high mortality in advanced patients. The epidemiological investigation in China indicated that the morbidity of ovarian cancer was approximately 52.1 per 100,000, and the mortality was 22.5 100,000 in 2015.[1] Lacking effective early detection methods and typical clinical symptoms, the proportion of advanced ovarian cancer was close to 70% at the time of diagnosis. Moreover, the 5-year survival rate of ovarian cancer fluctuated between 30% and 40% with limited improvement in the past 20 years, as cytoreductive operation followed by platinum-based chemotherapy was established as the standard therapy.[2] Currently, the effectiveness of initial therapy against ovarian cancer is 60% to 80%, however, as a prominent problem, the platinum resistance plays a negative role in subsequent therapy with depressingly poor prognosis.[3,4] In recent years, clinicians gradually concentrate on the roles of genetic and epigenetic backgrounds in the carcinogenesis, and relevant gene researches associating with platinum-resistance and ovarian cancer prognosis become important and essential.
Neurotrophic tyrosine receptor kinase 3 (NTRK3) is a member of neurotrophic tyrosine receptor kinase (NTRK) family with neurotrophin-3 (NT-3) as a ligand, which regulates the cell proliferation, differentiation, and apoptosis.[5–8] The functions of NTRK3 are ligand-dependent, transducing positive signals in the existence of the ligand, such as promoting growth and differentiation, but inducing apoptosis in the absence of the ligand.[3] NTRK3 also plays important roles in tumorigenesis, invasion, and metastasis.[9,10] Relevant studies have demonstrated that the fusion of erythroblastosis variant gene 6 (ETV-6) and NTRK3 promotes tumor formation and progression, such as secretory cancer of breast, gastrointestinal stromal tumors, and pediatric papillary thyroid carcinoma, by encoding a chimeric protein tyrosine kinase and playing oncogenic functions.[11–14] However, functions of NTRK3 may vary and the structural variants of NTRK3 also differ in different kinds of tumors. Recently, the missense mutation and methylation of NTRK3 have been identified in some tumors (colorectal cancer, breast cancer, lung cancer, and pancreatic cancer), which support the possible antioncogenic function of NTRK3.[9]
The possible mechanisms of NTRK3 are complex and the functions of it are divergent in different tumors. So far, the functions of NTRK3 and relevant structural variants in ovarian cancer have not been reported. Our study tried to evaluate the correlation between copy number variations (CNVs) of NTRK3 and the prognosis of ovarian cancer by array comparative genomic hybridization (CGH) and fluorescence in situ hybridization (FISH).
2. Materials and methods
2.1. Patients and tissue specimens
The tumor samples were collected from patients diagnosed with ovarian cancer for primary treatment who visited National Cancer Center/Cancer Hospital, Chinese Academy of Medical Sciences between January 1, 2008 and May 30, 2013. The study was approved by Institutional Review Broad of National Cancer Center /Cancer Hospital, Chinese Academy of Medical Sciences. Written informed consent was obtained from all patients.
Acquired tumor tissues of the primary debulking surgery were frozen in liquid nitrogen for at least 2 hours within 30 minutes after detachment from human body, and were placed with a subsequent long-term storage at –80°C. A total of 57 cases were included in the study and the inclusion criteria contained histologically confirmed epithelial ovarian cancer, FIGO (International Federation of Gynecology and Obstetrics) III–IV stages, more than 70% tumor tissue (confirmed by H&E staining), and primary cytoreductive surgery followed by 6 to 8 cycles of platinum-based chemotherapy.
2.2. Array CGH
Genomic deoxyribonucleic acids (DNAs) of 17 cases (8 cases with time to recurrence [TTR] >15 months and 9 cases with TTR ≤6 months) were extracted for the screening of copy number changes. DNA was digested with restriction enzyme and amplified with Exo-Klenow polymerases. The amplification product of control samples (commercially available genomic DNA extracted from women's peripheral blood) and study samples (genomic DNA extracted from tissues of ovarian cancer) were stained with Cy3 deoxyuridine triphosphate (dUTP) and Cy5-dUTP. After purification, concentrations of amplification product, incorporation rates, and fluorescence intensity values (Cy3-dUTP and Cy5-dUTP) were calculated. A comparable balance should be reached with differences less than 1.5-fold. Microarray Comparative Human Genome Hybridization (Agilent Oligo array CGH Hybridization Kit, Agilent Technologies, Santa Clara) was used for hybridization and Feature Extraction 9.5.3.1 Software combined with DNA analytics was used for the detection of signal intensity and amplification calculation with a quality control microarray. Gene chips after hybridization were scanned by Agilent Microarray Scanner and Agilent Scan Control with 5-μm resolution ratio. Array CGH Analytics 3.5.14 Software was used for homogenize fluorescence intensity values of probes for further analysis. A fold change with more than 1.5-fold was considered as statistically significant.
2.3. FISH
The interphase nucleus of ovarian cancer cells and lymphocytes deriving from peripheral blood of healthy control were prepared with low osmotic potassium chloride solution, colchicines, and fixation fluid (methanol: glacial acetic acid = 3:1) for preservation at 4°C. DNA in bacterial artificial chromosome (BAC) plasmid (the final concentration: 20 ng/μL; total: 8 μg) was extracted by means of wizard@ plus structure variation minipreps DNA purification system kits and was stained with Bio Nick TM Kits (GIBCO Corporation). The slides of interphase nuclei of ovarian cancer cells were made and interphase nuclei of lymphocytes on the same slide were used as control, using RNase (1 μL, 10 mg/mL) and sodium citrate buffer (100 μL, 1:100) for pretreatment. Hybridization was carried out after the degeneration of probes and disposal of slides. FISH images were obtained with the fluorescence microscope and the fluorescence signal observation of Cy3, spectrum-green and 4′, 6-diamidino-2-phenylindole (DAPI) was achieved by the usage of different filter. These signals were converted into the final colorful images with image processing software. The quality control criteria were more than 75% hybridization signals existing in normal nuclei (more than 100 cells) and no signal interference in extracellular backgrounds. Amplification was defined as the ratio of Cy3/Spectrum (RP11–91E10/15 chromosome centromere) or the signals of Cy3 and Spectrum ≥3 per cell in more than 30% of all cells.
2.4. Statistical analysis
TTR of all patients were retrieved by retrospective analysis according to electrical records. The relevant influencing factors, such as, ages, FIGO stages, differentiation, pathological types, residual tumors, neoadjuvant chemotherapy, were matched. Correlation between copy number variations and TTR was assessed by log-rank test. Spearman's rank correlation analysis, discriminant analysis and χ2 test were adopted to evaluate the predictive accuracy of CNVs of NTRK3 in 2 groups. The statistical product and service solutions (SPSS) 19.0 software was used for statistical analysis. Concordance between array CGH and FISH was tested by Spearman's rank correlation analysis and χ2 test. P <.05 was considered as statistically significant.
3. Results
3.1. Clinicopathological characteristics
A total of 57 cases were involved in the study and relevant clinicopathological characteristics are shown in Table 1. The therapy protocol was determined by gynecological examination and imaging examination. Among all patients, 31 cases received primary cytoreductive surgery. Others received 2 or 3 cycles of neoadjuvant chemotherapy (21 cases: paclitaxel plus cisplatin/carboplatin protocol; 5 cases: cyclophosphamide plus adriamycin plus cisplatin) with subsequent debulking operation. All of patients were administered with 6 cycles of platinum-based chemotherapy after operation.
Table 1.
Clinicopathological characteristics.
The median follow-up time was 34 months with recurrence in 49 cases till the last follow-up. Thirty-eight cases died and 19 cases were alive with or without recurrent foci. The median TTR was 9 months. The proportions of platinum-sensitive and platinum-resistant recurrences were, respectively, 61.4% (35/57) and 38.6% (22/57). The median overall survival time was 57 months. The 2-year and 5-year cumulative survival rates were, respectively, 80.9% and 42.4%.
3.2. CNVs of NTRK3 by array CGH
We compared the genetic backgrounds of platinum-sensitive group (8 cases) and platinum-resistant group (9 cases) regarding 6.0 as a threshold value. The outcomes of array CGH detected the copy number variations and the location of NTRK3 gene. The difference of copy number variations between platinum-sensitive group and platinum-resistant group was more than 1.5-fold with statistical significance. The amplification of NTRK3 was more common in platinum-sensitive group. The location and copy number variations of NTRK3 are demonstrated in Fig. 1. Considering the possible limitation of small samples receiving array CGH, we enlarged the number of patients to confirm the amplification status of NTRK3 in platinum-sensitive and platinum-resistant populations with FISH to evaluate the copy number variants.
Figure 1.
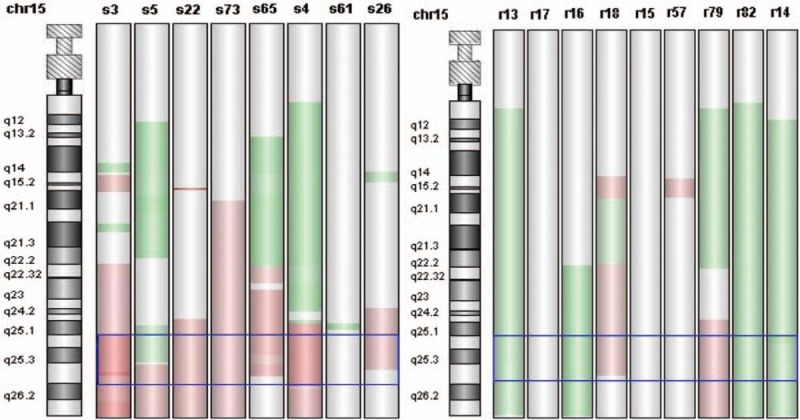
NTRK3 gene located in 15q25.3 and copy number variations of NTRK3 were demonstrated between platinum-sensitive and platinum-resistant groups. Pink = amplification; green = nonamplification; chr = chromosome; s = platinum-sensitive recurrence; r = platinum-resistant recurrence. NTRK3 = neurotrophic tyrosine receptor kinase 3.
3.3. CNVs of NTRK3 by FISH
Fifty-two cases were selected in FISH test, while the other 5 cases were not included due to 3-drug combination chemotherapy (cyclophosphamide plus adriamycin plus cisplatin). Within 52 patients, 11 cases were excluded because of not conforming to the signal standard (the detailed standard was shown in Methods section). Among the remaining 41 patients, 18 cases were manifested with amplification in 15q25.3 (probe: BAC clone RP11-91E10) and 23 cases showed negative results (Fig. 2). The median TTR of patients with NTRK3 amplification was 18 months, while the median TTR with NTRK3 amplification negative was only 5 months. The difference had statistical significance (P <.01), which is shown in Fig. 3.
Figure 2.
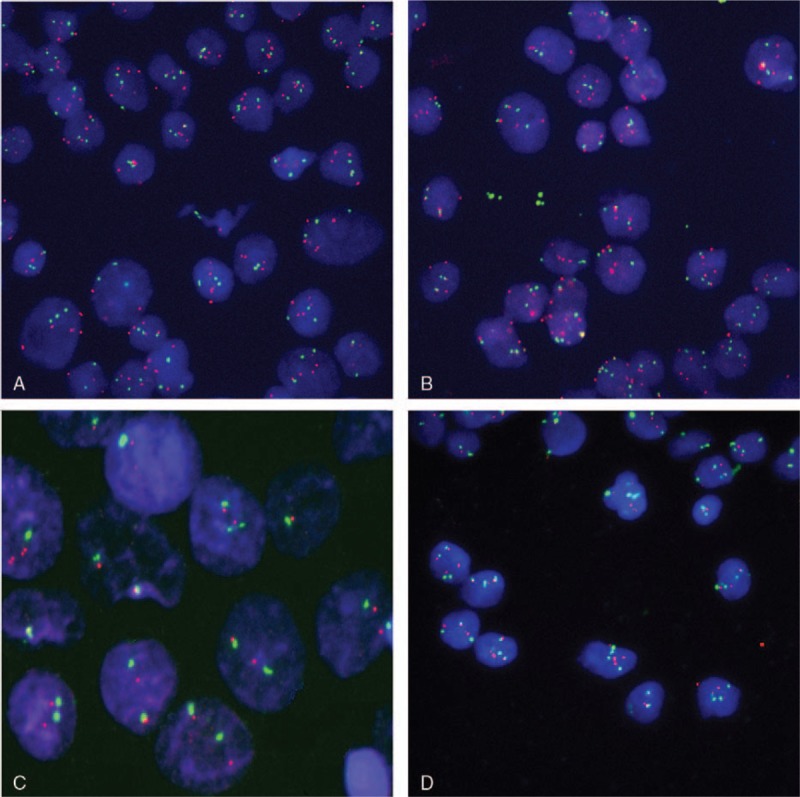
FISH images (NTRK3) of interphase (A, B, D 63 × , C 253 × ). Blue: interphase nuclei stained with DAPI; red: 15q25.3 labeled with Cy3; green: centrosome of 15 chromosome labeling with SpG; A and B: amplification; C: normal control; D: images without amplification. DAPI = 4′,6-diamidino-2-phenylindole, FISH = fluorescence in situ hybridization, NTRK3 = neurotrophic tyrosine receptor kinase 3.
Figure 3.

The median TTR of patients with NTRK3 amplification was 18 months, while the median TTR without NTRK3 amplification was only 5 months. Blue: amplification; green: nonamplification. NTRK3 = neurotrophic tyrosine receptor kinase 3, TTR = time to recurrence.
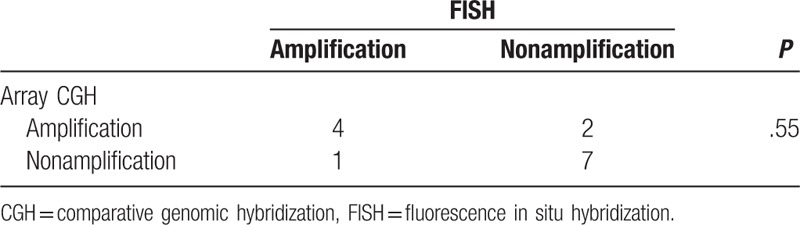
Among 17 cases received array CGH test, the results of 14 cases checked by FISH conformed to the standard. The analysis of 14 cases demonstrated that the copy number changes of NTRK3 were coincident in 11 cases. Accuracy rate of tests was 78.6% and there was no statistical difference between array CGH and FISH (Table 2).
Table 2.
Consistency analysis between FISH and array CGH.
3.4. CNVs of NTRK3 and prediction of chemotherapy sensitivity
The outcomes supported that amplification of NTRK3 was associated with promising response of initial paclitaxel plus platinum chemotherapy. The previous array CGH test also confirmed that amplification of NTRK3 was a favorable predictor. According to international guideline, platinum-resistant recurrence is defined as recurrence within 6 months; platinum-sensitive recurrence is prescribed as recurrence more than 6 months. The accuracy rate of NTRK3 to predict recurrence was 65.9% (27/41) and the copy number variations between 2 groups had a significant difference (Table 3).
Table 3.
Accuracy of NTRK3 amplification through FISH to predict platinum-sensitive and platinum-resistant recurrences.

4. Discussion
So far, NT-3 is the only known ligand of NTRK3 and regulates the functions of NTRK3 which is explained by the dependent receptor theory. The gene functions of NTRK3 are complex and relevant studies indicate that NTRK3 expression is not restricted to neural tissues, which is also found in hematopoietic cells, various types of epithelium and epidermis of skin. Signaling through the interaction between NT-3 and NTRK3 may induce pleiotropic functions in multiple organs or tissues.[15] Neither the copy number changes of NTRK3 nor the correlation between NTRK3 and ovarian cancer has been reported. The studies about NTRK3 in other tumors are also limited and the aberrance of NTRK3 includes ETV6-NTRK3 fusion, mutation of NTRK3, methylation of NTRK3, and so on. Recently some researchers focus on the possible functions of isoforms of NTRK3.[9,10,15,16]
ETV6-NTRK3 gene fusion was reported in malignant tumors, such as mammary analogue secretory carcinoma, hematopoietic malignancies, esophageal cancer, and congenital fibrosarcoma.[12–14] Wai et al[14] transformed ETV6-NTRK3 gene fusion in NIH3T3 cell lines and found that the fusion gene could encode a chimeric protein tyrosine kinase which was a strong oncogenic protein with transforming activity. The crucial kinase and pathway activated by ETV6-NTRK3 gene fusion were Ras-mitogen activated protein kinase (Ras-MAPK) pathways which were very important for cell proliferation and survival. Unbalance of growth and apoptosis lead to tumorigenesis.
Compared to normal tissues, NTRK3 was overexpressed in breast cancers and the phosphorylation of NTRK3 significantly increased in invasive human breast tumors, suggesting that overexpression of NTRK3 might contribute to the pathogenesis of human invasive breast carcinoma.[7,11] The research of Jin supported the oncogenic function of NTRK3, emphasizing the activation of mitogenic pathways through expression of NTRK3 in breast cancer cells. Therefore, the results conferred potent tumorigenic, invasive, and metastatic capacities, suggesting that NTRK3 might be directly associated with the genesis of human malignant tumor.[7]
However, contrary to the oncogenic role in breast cancer, NTRK3 was reported as tumor suppressor gene in colorectal cancer. Luo et al[9] found that NTRK3 expression without NT-3 could induce the apoptosis of tumor cell in vitro and in vivo, and this phenomenon could be reversed by reintroduction of NT-3. The mutation or methylation of NTRK3 observed in primary colorectal cancer could inactivate the function of NTRK3 and loss of NTRK3 led to the disorder of MAPK signaling pathway.[5,17,18] The above function of NTRK3 was remarkably different from the transforming activity of NTRK3 which was fused with EVT-6 in other cancers.
In our study, we confirmed the CNVs of NTRK3 in ovarian cancer through screening by array CGH and FISH. Compared to platinum-resistant population, amplification of NTRK3 was more common in the platinum-sensitive population. The TTR was significantly longer in NTRK3 amplification group than nonamplification group. There was an intimate correlation between the amplification of NTRK3 and chemotherapeutic response. Though there were no relevant articles revealing the potential impact of CNVs in ovarian cancer or other tumors, we detected important influence of overexpression of NTRK3 in medulloblastoma and melanoma.[19–22] Segal et al[19] tested NTRK3 expression in medulloblastoma and obtained similar conclusion through testing the expression of mRNA level and protein level. Kaplan–Meier analysis of 12 cases demonstrated that patients with overexpression of NTRK3 in mRNA level had significantly longer intervals without disease progression than those with low levels expression of NTRK3 (P <.01), and the overall survival was more favorable in the group of overexpression (P = .05). The studies of melanoma also showed that the prognosis was optimistic if the NTRK3 gene was overexpressed.[20] The function of tumor suppression of NTRK3 may be conditional, which depends on the existence of corresponding ligands. When the ligand existed, cellular differentiation and growth were promoted, however, the absence of ligand leaded to the cleavage of death-domain peptide which was intimately associated with cell apoptosis.[9,21,23]
Because of the relentless trajectory to eventual drug resistance, the therapy of ovarian cancer is clinically challenged by repeated recurrence. The copy number variations of NTRK3 preferably predict platinum-sensitive or platinum-resistant recurrence, indicating that the NTRK3 gene may play an important role in drug resistance. This may provide relevant target for treatment against platinum resistance of ovarian cancer.
There are some limitations of the study. The study about correlation between copy number variations of NTRK3 and relapse of ovarian cancer indicates that NTRK3 preferably predicts platinum-sensitive or platinum-resistant recurrence. The samples may be enlarged in the subsequent work in order to evaluate the incidence of CNVs of NTRK3 in larger population with ovarian cancer. The detailed aberrant information of NTRK3 in ovarian cancer (mutation, CNVs, methylation, or ETV6-NTRK3) and the status of transcription in mRNA/protein level still need a further study, which will provide more information about the functions of NKRK3 in ovarian cancer.
Acknowledgments
The authors thank Yang-Chun Sun for her intellectual support and they really appreciate the support of colleagues in the screening of electrical archives. They also thank Lu Wang for instructions about statistical analysis.
Footnotes
Abbreviations: CGH = comparative genomic hybridization, BAC = bacterial artificial chromosome, CNVs = copy number variations, DAPI = 4′,6-diamidino-2-phenylindole, DNA = deoxyribonucleic acid, ETV-6 = erythroblastosis variant gene 6, FIGO = International Federation of Gynecology and Obstetrics, FISH = fluorescence in situ hybridization, MAPK = mitogen activated protein kinase, NT-3 = neurotrophin-3, NTRK3 = neurotrophic tyrosine receptor kinase 3, SPSS = statistical product and service solutions, TTR = time to recurrence.
Funding: This work is funded by National Natural Science Foundation of China (81341076).
The authors have no conflicts of interest to disclose.
References
- [1].Chen W, Zheng R, Baade PD. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115–32. [DOI] [PubMed] [Google Scholar]
- [2].Gabra H. Introduction to managing patients with recurrent ovarian cancer. Eur J Cancer 2014;12:2–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ozols RF, Bundy BN, Greer BE, et al. Phase III trial of carboplatin and paclitaxel compared with cisplatin and paclitaxel in patients with optimally resected stage III ovarian cancer: a Gynecologic Oncology Group study. J Clin Oncol 2003;21:3194–200. [DOI] [PubMed] [Google Scholar]
- [4].Jayson GC, Kohn EC, Kitchener HC, et al. Ovarian cancer. Lancet 2014;384:1376–88. [DOI] [PubMed] [Google Scholar]
- [5].Bouzas-Rodriguez J, Cabrera JR, Delloye-Bourgeois C, et al. Neurotrophin-3 production promotes human neuroblastoma cell survival by inhibiting TrkC-induced apoptosis. J Clin Invest 2010;120:850–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ivanov SV, Panaccione A, Brown B, et al. TrkC signaling is activated in adenoid cystic carcinoma and requires NT-3 to stimulate invasive behavior. Oncogene 2013;32:3698–710. [DOI] [PubMed] [Google Scholar]
- [7].Jin W, Kim GM, Kim MS, et al. TrkC plays an essential role in breast tumor growth and metastasis. Carcinogenesis 2010;31:1939–47. [DOI] [PubMed] [Google Scholar]
- [8].Kim MS, Kim GM, Choi YJ, et al. TrkC promotes survival and growth of leukemia cells through Akt-mTOR-Dependent Up-Regulation of PLK-1 and Twist-1. Mol Cells 2013;36:177–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Luo Y, Andrew MK, Samornmas K, et al. NTRK3 is a potential tumor suppressor gene commonly inactivated by epigenetic mechanisms in colorectal cancer. PLoS Genet 2013;9:e1003552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wood LD, Calhoun ES, Silliman N, et al. Somatic mutations of GUCY2F, EPHA3, and NTRK3 in human cancers. Hum Mutat 2006;27:1060–1. [DOI] [PubMed] [Google Scholar]
- [11].Prasad ML, Vyas M, Horne MJ. NTRK fusion oncogenes in pediatric papillary thyroid carcinoma in northeast United States. Cancer 2016;122:1097–107. [DOI] [PubMed] [Google Scholar]
- [12].Laé M, Fréneaux P, Sastre-Garau X, et al. Secretory breast carcinomas with ETV6-NTRK3 fusion gene belong to the basal-like carcinoma spectrum. Mod Pathol 2009;22:291–8. [DOI] [PubMed] [Google Scholar]
- [13].Urano M, Nagao T, Miyabe S, et al. Characterization of mammary analogue secretory carcinoma of the salivary gland: discrimination from its mimics by the presence of the ETV6-NTRK3 translocation and novel surrogate markers. Hum Pathol 2015;46:94–103. [DOI] [PubMed] [Google Scholar]
- [14].Wai DH, Knezevich SR, Lucas T, et al. The ETV6-NTRK3 gene fusion encodes a chimeric protein tyrosine kinase. Oncogene 2000;19:906–15. [DOI] [PubMed] [Google Scholar]
- [15].Hisaoka M, Sheng WQ, Tanaka A, et al. Gene expression of TrkC (NTRK3) in human soft tissue tumours. J Pathol 2002;197:661–7. [DOI] [PubMed] [Google Scholar]
- [16].Bardelli A, Parsons DW, Silliman N. Mutational analysis of the tyrosine. Science 2003;300:949. [DOI] [PubMed] [Google Scholar]
- [17].Blasco-Gutierrez MJ. TrkC: a new predictive marker in breast cancer? Cancer Invest 2007;25:405–10. [DOI] [PubMed] [Google Scholar]
- [18].Genevois AL, Ichim G, Coissieux MM. Dependence receptor TrkC is a putative colon cancer tumor suppressor. Proc Natl Acad Sci U S A 2013;110:3017–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Segal RA, Goumnerova LC, Kwon YK, et al. Expression of neurotrophin receptor Trk-C. Proc Natl Acad Sci 1994;91:12867–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Xu X, Tahan SR, Pasha TL, et al. Expression of neurotrophin receptor Trk-C in nevi and melanomas. J Cutan Pathol 2003;30:318–22. [DOI] [PubMed] [Google Scholar]
- [21].Goldschneider D, Mehlen P. Dependence receptors: a new paradigm in cell signaling and cancer therapy. Oncogene 2010;29:1865–82. [DOI] [PubMed] [Google Scholar]
- [22].Yamashiro DJ, Liu XG, Lee CP, et al. Expression and function of Trk-C in favourable human. Eur J Cancer 1997;33:2054–7. [DOI] [PubMed] [Google Scholar]
- [23].Blume-Jensen P, Hunter T. Oncogenic kinase signaling. Nature 2001;411:355–65. [DOI] [PubMed] [Google Scholar]


