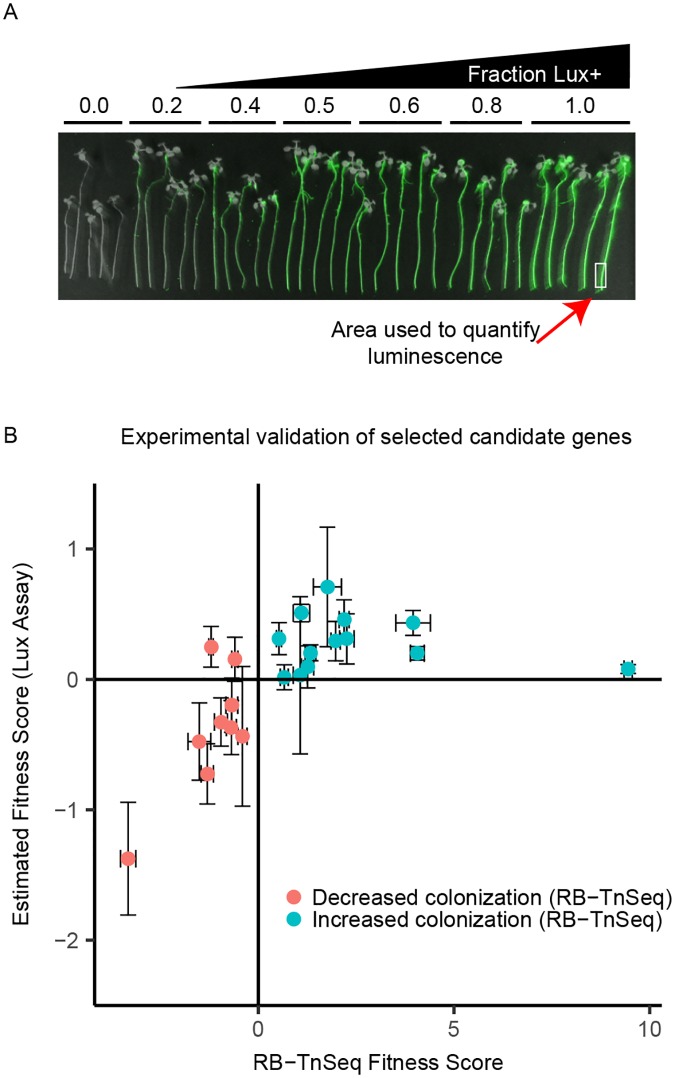
Fig 3. Selected candidates are validated using secondary luciferase-based screen.
Twenty-two mutant strains were retrieved from an arrayed clone library for further validation. These strains were competed against an engineered wild-type (WT) P. simiae strain that produces luciferase. (A) False-color image of competition between the nonengineered WT WCS417r strain versus the luciferase-producing (Lux+) strain. Each group of 5 roots represents a different ratio of Lux+ to WT (0.0–1.0). Luminescence intensity is false colored in green. White rectangle indicates approximate region of the root tip used to measure luciferase activity. (B) All 22 insertion mutant strains (described in S1 Data) were competed with the Lux+ on roots and empty phytagel/mesh plates after inoculation as a 1:1 mixture. A ratio of each mutant strain to the Lux+ strain on the roots was estimated by interpolating the luminescence intensity of the root tip onto a standard curve (S5 Fig, S1 Data). The estimated fitness score (Lux+ colonization index [CI] in S1 Data) was then derived from the log-transformed root ratio minus the log-transformed mesh (no root final [NRF]) ratio (see S1 Data). Error bars are ± standard error (n = 3 biological replicates for y-axis, n = 15 for x-axis). Abbreviation: RB-TnSeq, randomly barcoded transposon mutagenesis sequencing.