Abstract
Background
Prostate cancer (CP) cells differ from their normal counterpart in gene expression. Genes encoding secreted or extracellular proteins with increased expression in CP may serve as potential biomarkers. For their detection and quantification, assays based on monoclonal antibodies are best suited for development in a clinical setting. One approach to obtain antibodies is to use recombinant proteins as immunogen. However, the synthesis of recombinant protein for each identified candidate is time-consuming and expensive. It is also not practical to generate high quality antibodies to all identified candidates individually. Furthermore, non-native forms (e.g., recombinant) of proteins may not always lead to useful antibodies. Our approach was to purify a subset of proteins from CP tissue specimens for use as immunogen.
Methods
In the present investigation, ten cancer specimens obtained from cases scored Gleason 3+3, 3+4 and 4+3 were digested by collagenase to single cells in serum-free tissue culture media. Cells were pelleted after collagenase digestion, and the cell-free supernatant from each specimen was pooled and used for isolation of proteins in the 10–30 kDa molecular weight range using a combination of sonication, dialysis and Amicon ultrafiltration. Western blotting and mass spectrometry (MS) proteomics were performed to identify the proteins in the selected size fraction.
Results
The presence of cancer-specific anterior gradient 2 (AGR2) and absence of prostate-specific antigen (PSA)/KLK3 were confirmed by Western blotting. Proteomics also detected AGR2 among many other proteins, some outside the selected molecular weight range, as well.
Conclusions
Using this approach, the potentially harmful (to the mouse host) exogenously added collagenase was removed as well as other abundant prostatic proteins like ACPP/PAP and AZGP1 to preclude the generation of antibodies against these species. The paper presents an optimized scheme for convenient and rapid isolation of native proteins in any desired size range with minor modifications.
Keywords: Prostate cancer proteins (CP proteins), purification of 10–30 kDa proteins, anterior gradient 2 (AGR2), cancer biomarkers, proteomic analysis
Introduction
Other than skin cancer, prostate cancer (CP) is the most common cancer in American men. According to the American Cancer Society’s estimate, 180,890 new cases of CP will be diagnosed and 26,120 men will die of the disease in 2016. About 1 in 7 men will be diagnosed with CP during his lifetime (1). Nearly two-thirds of the cancer is diagnosed in men aged 65 or older. CP is the second leading cause of cancer death in American men, but most diagnosed men do not die from it. In fact, more than 2.9 million men in the United States who have been diagnosed with CP at some point are alive today. The current biomarker of elevated serum prostate-specific antigen (PSA) is a flawed test for early detection as many men with abnormal serum PSA turn out not to have cancer upon biopsy.
Researchers are working on strategies to improve molecular diagnosis. One approach is to assay for specific characteristics of PSA, or to detect PSA variants more specific to cancer (2). Another approach is to develop new tests based on other tumor markers. We used comparative analysis between the transcriptomes of isolated CD26+ cancer cells and CD26+ normal luminal cells to identify genes up-regulated in cancer (3,4). Genes that were found to be overexpressed by ≥8-fold, and to encode secreted or extracellular proteins were selected as biomarker candidates for assay development. Similarly, comparative analysis of isolated CD90+ cancer-associated stromal cells and CD49a+ benign tissue stromal cells has identified additional secreted protein candidates (5,6).
Anterior gradient 2 (AGR2; 19 kDa), breast cancer membrane protein 11/AGR3 (BCMP11; 19 kDa), cysteine-rich secretory protein-3 (CRISP3; 28 kDa), thymocyte differentiation antigen 1/CD90 (THY1; ~27 kDa) are examples of proteins secreted or released from prostate tumors. As such, these proteins can all likely be detected and measured in body fluids. Furthermore, array signals indicate that AGR2 is a moderately abundant protein while the others are less (3,7). In order to obtain antibodies that can recognize AGR2 and others, one needs to isolate these antigens preferably in their native form for mouse immunization.
The present study concerns with the development of a procedure to isolate cancer secreted/extracellular proteins such as AGR2 from media in which CP tissue was digested by collagenase (8). This media contained proteins synthesized by cell types of the tumors. Since a number of biomarker candidates are calculated to be in the 10–30 kDa size range, protein separation based on size fractionation was employed. The rationale behind the choice of 30 kDa as the upper bound was to ensure that monoclonal antibodies would not be raised against the abundant PSA (34 kDa) and other prostatic proteins such as zinc α2-glycoprotein (AZGP1, 43 kDa) and prostatic acid phosphatase (ACPP, 52 kDa), and to remove collagenase type I (68 kDa) from being present in the protein preparation. Another abundant species, β-microseminoprotein (MSMB/prostate secretory protein of 94 amino acids; 13 kDa), shows much reduced expression in tumors (3). The choice of 10 kDa as the lower bound was to remove impurities and small molecules such as peptide fragments.
The tissue digestion media used in this study was media supernatant pooled from ten tumor tissue specimens of Gleason 3+3, 3+4, and 4+3 scores (and one of 4+5), which would contain proteins synthesized by both pattern 3 and pattern 4 tumors. Proteins in the desired molecular weight range were purified by a scheme involving sonication, dialysis and ultrafiltration. Western blotting analysis was done to ascertain whether the resultant purified product was, for example, positive for AGR2 and negative for PSA. Mass spectrometry (MS)-based proteomics was also used to profile the protein composition of the purified fraction to gauge proteomic complexity.
Methods
Ethical statement
Human prostate tissue specimens were obtained from ten radical prostatectomy patients operated by surgeons in the Department of Urology with informed and written consent from all participants as per the guidelines approved by the University of Washington Institutional Review Board. Frozen sections were histologically assessed to confirm that the specimens were enriched for cancer. The pathology characteristics of the collected tumors were as follows: 05-081CP G3+3; 05-020CP G3+4; 05-036CP G3+4; 05-125CP G3+4; 05-056CP G3+4; 05-131CP G3+4; 05-081CP G3+3; 03-169CP G3+3; 05-154CP G4+5; 04-176CP G4+3 (G = Gleason score).
Tissue specimens and tissue digestion
Each CP tissue sample weighing at least 0.1 g, was rinsed with Hanks balanced salt solution (HBSS) and minced for enzymatic digestion overnight at room temperature with 0.2% collagenase type I (Invitrogen, Carlsbad, CA, USA) in serum-free RPMI1640 media on a magnetic stirrer. The resultant cell suspension was filtered with a 70-μm Falcon cell strainer, diluted with an equal volume of HBSS, and aspirated with an 18-gauge needle. The suspension was centrifuged at 1,000 rpm for 15 min to collect the supernatant. These media preparations were screened by Western blotting analysis to show low levels of TIMP1 and high levels of CD90 compared with normal tissue (NP) or benign prostatic hyperplasia (BPH) samples digested by collagenase (5). Tumor loses TIMP1 expression (8).
Purification of proteins in the 10–30 kDa range
To 22.5 mL of pooled CP samples, the media supernatant was made to contain 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM benzamidine, 10 μM leupeptin, 1 μM pepstatin A, 1 mM EDTA, 1% Triton-X-100 and 15 mM β-mercaptoethanol. The sample was stirred for 1 h at 4 °C. The pH was adjusted to 5.5, and the solution was stirred for another 1 h. The sample was then sonicated three times for 2 min each, and centrifuged at 40,000 rpm for 30 min. The supernatant was then passed through a 0.22-micron filter, and fractionated using a 30-kDa MWCO Millipore filter (Millipore, Billerica, MA, USA) at 1,000 rpm for 30 min. The flow-through was then fractionated using a 10-kDa MWCO Millipore filter at 4,000 rpm for 30 min. The resultant product after these filtrations was concentrated to 1 mL. One mL MES buffer (25 mM MES, pH 5.5, 150 mM NaCl, 1 mM EDTA, 50 mM β-mercaptoethanol) was then added. The retentate was centrifuged at 4,000 rpm to about 1 mL volume. This preparation was desalted by placing in 3-kDa dialysis tubing overnight against 1 mM MES, pH 5.5.
SDS-PAGE and Western blot analysis
Protein concentration of the purified CP sample was measured by Bradford Assay (BioRad, Hercules, CA, USA). For gel analysis, loading buffer containing 0.1 M DTT was added to the amount of purified CP sample containing ~60 μg protein, heated to 70 °C for 10 min, electrophoresed on 4–20% gradient SDS-polyacrylamide gel (BioRad), and electrotransferred to PVDF membrane (Hybond-P, Amersham). Blotting was ascertained by visualization of protein bands on the membrane by Ponçeau S (Sigma, St. Louis, MO, USA). Afterwards, the stain was completely removed by repeated washing with triple distilled water. The membrane was immersed in 5% nonfat dry milk-PBS-Tween for 30 min, and probed with AGR2 antibody (1:2,000; clone 1C3, Abnova, Taiwan), or PSA antibody (1:1,000; clone A67-B/E3, Santa Cruz Biotechnology, Dallas, TX, USA) for 60 min, followed by horseradish peroxidase conjugated anti-mouse IgG (1:5,000; Vector, Burlingame, CA, USA). After washing, the membrane was incubated with Luminol (Santa Cruz Biotechnology), and the immunoreactive bands were visualized using Biomax MR light film (Kodak, Rochester, NY, USA).
Proteomic analysis using liquid chromatography-tandem mass spectrometry (LC-MS/MS)
The purified protein sample was further concentrated using Amicon Ultra-15 centrifugal filter MWCO3000 (Millipore). Protein concentration was determined by BCA Protein Assay (Thermo Scientific, Rockford, IL, USA). An aliquot of the concentrated preparation containing 10 μg protein was denatured in 50% trifluoroethanol at 60 °C for 2 h with gentle shaking (9), and then reduced with 2 mM DTT at 37 °C for 1 h. The sample was diluted 5-fold with 50 mM NH4HCO3 before digestion with trypsin (trypsin:protein 1:50 w/w) at 37 °C for 3 h. After speed-vac, the resultant tryptic peptides were reconstituted in 50 mM NH4HCO3 for LC-MS analysis.
LC-MS/MS was performed as previously described (10). Briefly, ~1 μg peptides were loaded onto a 65-cm-long, 75-μm-i.d. reversed-phase capillary column packed in house with 3 μm Jupiter C18 particles (Phenomenex, Torrance, CA, USA). The mobile phases consisted of solution A (0.1% formic acid in water) and solution B (0.1% formic acid in acetonitrile). An exponential gradient starting with 100% of mobile phase A to 60% of mobile phase B over the course of 100 min was employed with a flow-rate of ~500 NL/min. Eluted peptides were ionized via a nanoelectrospray ionization interface manufactured in house and analyzed on an LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA). The instrument was operated in data-dependent mode with m/z ranging from 400–2,000, in which a full MS scan was followed by ten MS/MS scans.
MS/MS data was analyzed by SEQUEST-based database searching against the Human International Protein Index (IPI) database (version 3.54). Searching parameters were: 3 Da tolerance for precursor ion masses and 1 Da for fragment ion masses with no enzyme restraint and a maximum of three missed tryptic cleavages. Filtering criteria (11) were applied for peptide identifications to achieve a <5% false discovery rate at the unique peptide level based on a reversed-database searching methodology (12).
Results
Purification of the 10–30 kDa fraction of CP tissue digests
Protein concentration of the purified CP sample was found to be at 3.2 mg/mL. Figure 1 shows the SDS-PAGE/Coomassie profile of the purified 10–30 kDa fraction. There was a good removal of proteins having molecular weights above 30 kDa by the purification scheme. At the same time, there were little 10–30 kDa proteins in the excluded fraction. The banding pattern also indicated minimal protein degradation.
Figure 1.
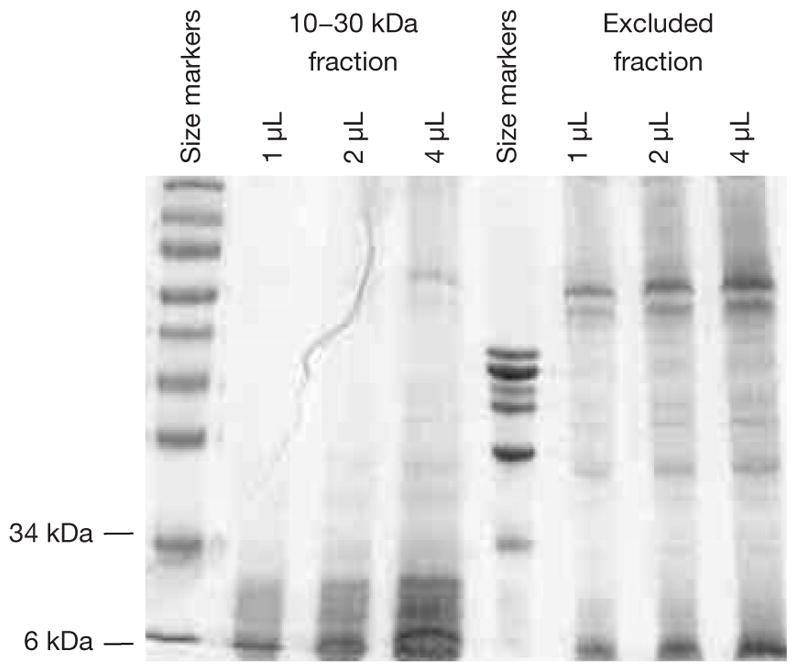
SDS PAGE-Coomassie analysis of purified 10–30 kDa fraction from prostate cancer (CP) tissue digest on a 4–20% gradient gel. For comparison, the excluded fraction was also analyzed.
Detection of AGR2 in the 10–30 kDa fraction
Figure 2 shows the presence of AGR2 (19 kDa) in the purified 10–30 kDa CP preparation and not in the excluded fraction. The Western blot also shows that this protein was not expressed by non-cancer (e.g., samples 05-091NP and 03-169BPH). There was an estimated loss of about 50% in the amount of AGR2 after purification (compare lanes labeled unfractionated CP and 10–30 kDa fraction, Figure 2). A fainter band lower than AGR2 may represent AGR3 (166 aa), which has 65% sequence homology with AGR2 (175 aa).
Figure 2.
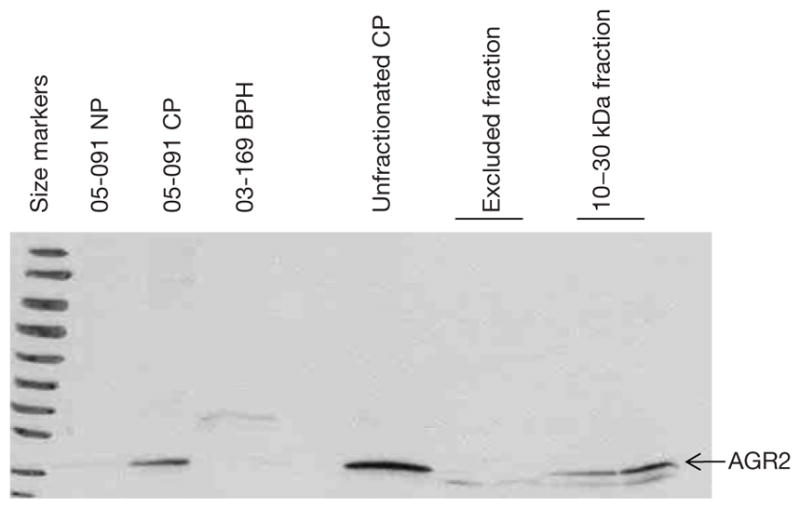
Western blot analysis of 10–30 kDa fraction from prostate cancer (CP) tissue digests. The membrane was probed with anti-anterior gradient 2 (AGR2) and the fraction was found to be AGR2 positive.
Absence of PSA in the 10–30 kDa fraction
The purified 10–30 kDa fraction was found to be negative for PSA as the unfractionated preparation was positive (Figure 3). On the blot, PSA was detected in matched 05-091CP and 05-091NP, as well as in 03-169BPH. Thus, the purification scheme was effective in eliminating an abundant protein species just 4 kDa larger than the upper size cutoff point.
Figure 3.
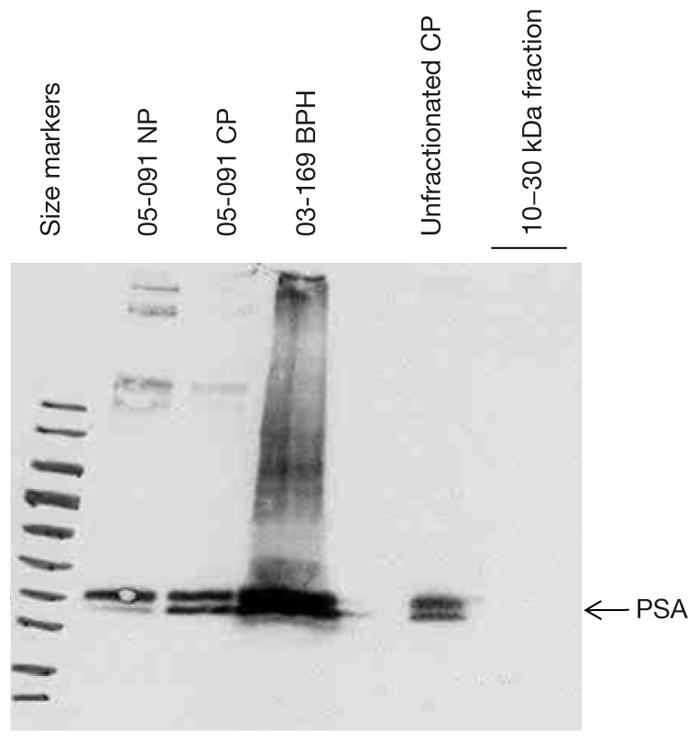
Western blot analysis of 10–30 kDa fraction from prostate cancer (CP) tissue digests. The membrane was probed with anti-prostate-specific antigen (PSA) and the fraction was found to be PSA negative.
MS analysis of purified CP fraction
The purified 10–30 kDa was analyzed using LC-MS/MS, and the proteins and peptides identified are included in Table S1. AGR2 was confidently identified by seven different peptides (Table 1). Previously, using unfractionated CP tissue digest media LC-MS/MS was unable to confidently detect AGR2 (unpublished) most likely due to its relatively low abundance or physical properties. On the other hand, the exogenously added (abundant) collagenase was not seen among the 252 proteins identified with two or more unique peptides. Although PSA was undetected by Western blotting, the protein was still identified by MS with seven spectral counts, but as evidence of a relatively low-abundance protein in the fraction. Table 2 lists the top dozen abundant proteins identified by LC-MS/MS using spectral counts as a measure of abundance (13). Among them are several large cytoskeletal components FLNA, COL6A3, FLNB, MYH11, VCL, etc. In fact, only 51% (290/574) of the proteins had calculated molecular weights in the 10–30 kDa range (and 1.6% below 10 kDa). Many of the proteins belonged to the cytoplasmic proteome and were not necessarily secreted.
Table 1.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis of the purified 10–30 kDa fraction. Anterior gradient 2 (AGR2) was identified by seven different tryptic peptides
| Peptide sequence | Protein name | Charge state | Xcorr of max | DelCN20f max | PPM of min | Cleavage type | Peptide sequence of count |
|---|---|---|---|---|---|---|---|
| K.LAEQFVLLNLVYETTDK.H | IPI:IPI00007427.2 | 2 | 5.3495 | 0.4284 | −9.38347 | 2 | 4 |
| L.YAYEPADTALLLDNMK.K | IPI:IPI00007427.2 | 2 | 2.7295 | 0.1342 | −3.50132 | 1 | 1 |
| R.GWGDQLIWTQTYEEALYK.S | IPI:IPI00007427.2 | 2 | 4.1530 | 0.3242 | −4.62505 | 2 | 2 |
| R.IMFVDPSLTVR.A | IPI:IPI00007427.2 | 2 | 3.0653 | 0.1520 | −6.01084 | 2 | 2 |
| R.LYAYEPADTALLLDNMK.K | IPI:IPI00007427.2 | 2 | 4.7862 | 0.4556 | −5.99701 | 2 | 3 |
| Y.AYEPADTALLLDNMK.K | IPI:IPI00007427.2 | 2 | 3.6434 | 0.3572 | −6.41511 | 1 | 2 |
| T.VQEESEEEEVDETGVEVK.D | IPI:IPI00007471.2 | 2 | 6.8026 | 0.3457 | −2.34021 | 1 | 2 |
Table 2.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) identification of the 12 most abundant proteins based on spectral counts
| Protein name | Peptide count | Total count | Description | Locus | MW |
|---|---|---|---|---|---|
| IPI:IPI00302592.2 | 56 | 148 | Isoform 2 of filamin-A | FLNA | 280018 |
| IPI:IPI00022434.4 | 19 | 81 | Putative uncharacterized protein ALB | ALB | 71704 |
| IPI:IPI00216138.6 | 15 | 79 | Transgelin | TAGLN | 22611 |
| IPI:IPI00025252.1 | 38 | 72 | Protein disulfide-isomerase A3 | PDIA3 | 56782 |
| IPI:IPI00021812.2 | 48 | 67 | Neuroblast differentiation-associated protein AHNAK | AHNAK | 629101 |
| IPI:IPI00022200.3 | 33 | 58 | Isoform 1 of collagen alpha-3(VI) chain | COL6A3 | 343665 |
| IPI:IPI00027230.3 | 25 | 51 | Endoplasmin | HSP90B1 | 92469 |
| IPI:IPI00289334.1 | 28 | 47 | Isoform 1 of filamin-B | FLNB | 278195 |
| IPI:IPI00179330.6 | 5 | 42 | Ubiquitin and ribosomal protein S27A precursor | UBC | 17965 |
| IPI:IPI00020501.1 | 20 | 41 | Myosin-11 | MYH11 | 227339 |
| IPI:IPI00010796.1 | 22 | 36 | Protein disulfide-isomerase | P4HB | 57116 |
| IPI:IPI00291175.7 | 16 | 32 | Isoform 1 of vinculin | VCL | 116722 |
Discussion
With protein biomarker discovery, reagents such as high quality antibodies are needed for clinical assays to measure these markers. Purified antigens are the optimal immunogens for monoclonal antibody production. However, the purification of multiple proteins from biosamples is extremely challenging and not cost-effective. The purification of 10–30 kDa protein species from pooled CP samples represents our attempt to obtain suitable materials for subsequent antibody production. Simple modifications can be done to the protocol to obtain any size fractions containing other informative biomarkers. Our results showed that this procedure is effective with exclusion of proteins outside the size range, as probed by Western blotting, while retaining the targeted proteins with acceptable losses. Importantly, the proteins are present in their native configuration with proper post-translational modifications intact. Antibodies resulting after mouse immunization would likely recognize specifically these protein analytes in human biospecimens. The laborious part would involve extensive screening of the hybridomas for the desired clones against particular antigens as the number of protein species in the 10–30 kDa CP is unknown (at least 574 based on the MS data). Removal of the abundant prostatic proteins ensures against generation of many unwanted antibodies. One drawback of this approach is that proteins in lower abundance would have fewer resultant clones, and screening for them would be hard. The first screen would compare immunoreactivity on frozen tissue sections of CP vs. NP. Those that show no reactivity on NP would be used for immunoprecipitation of CP digestion media or media of CP cell lines (7), and the protein(s) captured would be identified by MS. Most likely, these antibodies will not work well on Western blotting.
Using the more sensitive MS analysis, the 10–30 kDa fraction was shown to still contain nearly 50% protein species with molecular weights outside the selected range. These could result from degradation during the overnight tissue digestion. Frequently, some tumor tissue samples may contain necrotic areas not visible grossly. The cytoplasmic proteins then, most likely, resulted from breakdown of cells. This creates a potentially large obstacle for our approach as cytoskeletal proteins are some of the most immunogenic molecules.
An alternative to using CP tissue media is media supernatant of cultured cancer cell lines (and cancer-associated stromal cells) as well as that of xenograft tissue digest. Perhaps then, the problem of cell breakdown could be minimized. While no single cell line produces all the proteins found in CP (3), a panel of available cell lines and xenografts could produce most of these proteins. For example, AGR2 is made by CL1, CRISP3 by LuCaP 35, CEACAM5, BCMP11 by LuCaP 49 [results from dataset queries (4)]. In this way, we are not limited by the availability of ample material for immunization.
Our protein purification scheme has been optimized by including relatively fewer and simpler steps to obtain a good yield. It is convenient and efficient. The more common approach of using recombinant proteins as immunogens suffers from the lack of proper configuration when made in a bacterial or yeast host system, or low yield and high cost when made in mammalian cultured cells. The recombinantly produced proteins would still require purification. Nevertheless, useful monoclonal antibodies against AGR2 have been obtained by us using bacterially made recombinant AGR2. The selected antibody clones were able to recognize the recombinant protein and native AGR2 in CP (and not NP) digestion media, media supernatant of cultured CL1 cells, and in urine of patients (7). This could be due to the relatively small size of AGR2 and its lack of extensive glycosylation (small molecular weight difference between calculated size from amino acid composition and size estimated in SDS-PAGE).
The presence of stromal cell-derived proteins in the CP fraction shows that these could also be used for tumor detection. CD90 was previously detected by global proteomics in unfractionated CP tissue media (6,8). CD90 expression is increased in the tumor stroma compared with NP stroma, and can be measured in urine of patients (6). In addition, a number of other secreted CP stromal proteins have been identified. Thus, a CP tissue sample would contain proteins derived from both the cancer epithelial component and the tumor stromal component.
Conclusions
This work reports a method to selectively isolate native proteins in a particular molecular weight range using tissue digestion media. This process essentially lowers the proteomic complexity of the biosample that could be used to identify biomarker candidates, and for mouse immunization.
Supplementary Material
Acknowledgments
The authors thank Charles Luo and Michael Regnier in the Department of Bioengineering, Institute for Stem Cell and Regenerative Medicine, University of Washington, Seattle, USA, for their expertise in developing the purification protocol. Proteomics experiments were performed in the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the DoE and located at Pacific Northwest National Laboratory, which is operated by Battelle Memorial Institute for the DoE under Contract DE-AC05-76RL0 1830.
Funding: This work was supported by NCI-EDRN grant CA111244 and NIH P41GM103493.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The study was approved by the University of Washington Institutional Review Board and written informed consent was obtained from all patients.
Contributions: (I) Conception and design: AY Liu; (II) Administrative support: AY Liu; (III) Provision of study materials or patients: R Ahmad; (IV) Collection and assembly of data: R Ahmad; (V) Data analysis and interpretation: R Ahmad, CD Nicora, AK Shukla, RD Smith, WJ Qian; (VI) Manuscript writing: All authors; (VII) Final approval of manuscript: All authors.
References
- 1.American Cancer Society. Prostate Cancer. Available online: http://www.cancer.org/cancer/prostatecancer/detailedguide/index.
- 2.Sokoll LJ, Sanda MG, Feng Z, et al. A prospective, multicenter, National Cancer Institute Early Detection Research Network study of [−2]proPSA: improving prostate cancer detection and correlating with cancer aggressiveness. Cancer Epidemiol Biomarkers Prev. 2010;19:1193–200. doi: 10.1158/1055-9965.EPI-10-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pascal LE, Vêncio RZ, Page LS, et al. Gene expression relationship between prostate cancer cells of Gleason 3, 4 and normal epithelial cells as revealed by cell type-specific transcriptomes. BMC Cancer. 2009;9:452. doi: 10.1186/1471-2407-9-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pascal LE, Deutsch EW, Campbell DS, et al. The urologic epithelial stem cell database (UESC) - a web tool for cell type-specific gene expression and immunohistochemistry images of the prostate and bladder. BMC Urol. 2007;7:19. doi: 10.1186/1471-2490-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pascal LE, Goo YA, Vêncio RZ, et al. Gene expression down-regulation in CD90+ prostate tumor-associated stromal cells involves potential organ-specific genes. BMC Cancer. 2009;9:317. doi: 10.1186/1471-2407-9-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.True LD, Zhang H, Ye M, et al. CD90/THY1 is overexpressed in prostate cancer-associated fibroblasts and could serve as a cancer biomarker. Mod Pathol. 2010;23:1346–56. doi: 10.1038/modpathol.2010.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wayner EA, Quek SI, Ahmad R, et al. Development of an ELISA to detect the secreted prostate cancer biomarker AGR2 in voided urine. Prostate. 2012;72:1023–34. doi: 10.1002/pros.21508. [DOI] [PubMed] [Google Scholar]
- 8.Liu AY, Zhang H, Sorensen CM, et al. Analysis of prostate cancer by proteomics using tissue specimens. J Urol. 2005;173:73–8. doi: 10.1097/01.ju.0000146543.33543.a3. [DOI] [PubMed] [Google Scholar]
- 9.Wang H, Qian WJ, Mottaz HM, et al. Development and evaluation of a micro- and nanoscale proteomic sample preparation method. J Proteome Res. 2005;4:2397–403. doi: 10.1021/pr050160f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L, Aryal UK, Dai Z, et al. Mapping N-linked glycosylation sites in the secretome and whole cells of Aspergillus niger using hydrazide chemistry and mass spectrometry. J Proteome Res. 2012;11:143–56. doi: 10.1021/pr200916k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qian WJ, Kaleta DT, Petritis BO, et al. Enhanced detection of low abundance human plasma proteins using a tandem IgY12-SuperMix immunoaffinity separation strategy. Mol Cell Proteomics. 2008;7:1963–73. doi: 10.1074/mcp.M800008-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qian WJ, Liu T, Monroe ME, et al. Probability-based evaluation of peptide and protein identifications from tandem mass spectrometry and SEQUEST analysis: the human proteome. J Proteome Res. 2005;4:53–62. doi: 10.1021/pr0498638. [DOI] [PubMed] [Google Scholar]
- 13.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


