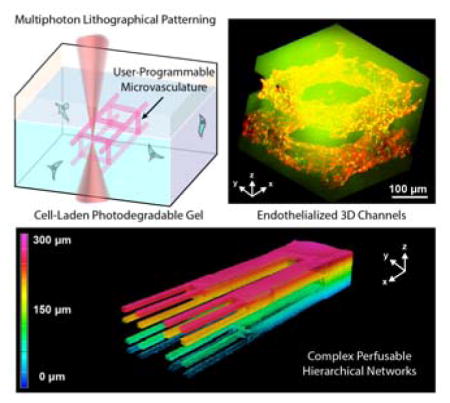
Abstract
A photodegradable material-based approach to generate endothelialized 3D vascular networks within cell-laden hydrogel biomaterials is introduced. Exploiting multiphoton lithography, microchannel networks spanning nearly all size scales of native human vasculature are readily generated with unprecedented user-defined 4D control. Intraluminal channel architectures are fully customizable, providing new opportunities for next-generation microfluidics and directed cell function.
Keywords: biomaterials, tissue engineering, vascular engineering, photodegradation, hydrogel
Graphical Abstract
A photodegradable material-based approach to generate endothelialized 3D vascular networks within cell-laden hydrogel biomaterials is introduced. Exploiting multiphoton lithography, microchannel networks spanning size scales of nearly all native human vasculature are readily generated with unprecedented user-defined 4D control. Intraluminal channel architectures are fully customizable, providing new opportunities for next-generation microfluidics and directed cell function.
Main Text
Vascularity is an integral aspect of all organ systems as it facilitates transport of oxygen, nutrients, hormones, electrolytes, and cells to and from every tissue within the body.[1] This vast interconnected network helps ensure temperature, pH, oxygen, glucose, and salt homeostasis, permits organ-to-organ hormonal communication, and enables rapid humoral and cell-mediated immune responses through convective transport of blood constituents. As all cells require nutrients and oxygen throughout maturation, blood vessels develop early in embryogenesis and synchronously with the organ systems they support.[2] During this time, vessels constantly change their shape and size to adapt to tissue requirements; adult vascular geometry is therefore four-dimensionally complex and ordered, spanning many scales of size.[3] Hierarchical organization of large arteries, to smaller arterioles, to the smallest capillaries dictates blood flow distribution and minimizes turbulence while the large size range allows for varying function. Centimeter-sized vessels including the aorta permit large convective volume flow while micron-sized capillaries support mass transport through diffusion. Bridging these vessels of differing sizes and structure is a shared and continuous tunica intima lined with endothelial cells, which serves functionally to prevent thrombosis, interpret changes in blood flow and composition, and modulate vascular tone.[4] This complex, hierarchical, and endothelial-lined 3D vascular network is critical for all bodily functions and should not be ignored when developing pathological models or engineering synthetic tissues.
As blood vessels are essential for tissue function, many synthetic approaches to generate macro- and microvasculature in vitro have been attempted, largely through additive- or subtractive-based methodologies.[5] Additive techniques generate vascular constructs from the bottom up, either through stacking of 2D constructs[6] or layer-by-layer 3D printing,[7–9] where void spaces intentionally left during manufacturing define construct vascularity. Though these approaches have proven successful in generating perfusable vessels of varying geometry, studying endothelial cell function,[10] and modeling vascular pathology,[11] limited 3D control has been afforded. Additive strategies further suffer from a frequent lack of cytocompatibility during material casting, preventing vessel creation in the presence of live cells, as well as an inability to generate channels with diameters smaller than a few hundred microns.[12]
To circumvent the complications of additive manufacturing-based vessel engineering, researchers have turned to subtractive techniques in which a bulk material is generated first followed by vascular void space creation.[13] Of note are recent efforts towards creating complex 3D networks using sacrificial lattices. In their seminal work, Miller et al encapsulated 3D-printed sugar glass in bioactive matrices, and then dissolved the lattice to reveal patterned interconnected channel networks.[14] Though this approach enables rapid vessel generation within cell-laden materials, concerns remain over cellular hyperglycemic response following hydration. Moreover, because the sugar network itself is generated through additive manufacturing techniques, the strategy exhibits the same limitations with respect to vessel 3D control, confining features to those >150 μm in size and with limited overhang.
Here and in literature, it has been hypothesized that vascular networks featuring capillary-sized microvessels could be generated using light-mediated subtractive manufacturing techniques.[15] Rastered control of a focused laser within biomaterials enables user-specified vascular creation of virtually any shape.[16,17] Utilization of a pulsed laser source, whereby multiple photons must be absorbed in rapid succession to initiate material dissolution, enables micron-scale control over vessel structure in all three spatial dimensions and provides access to complex and hierarchical networks similar to those found in vivo. Vessel geometry can be altered dynamically at any time based on subsequent light exposure, thereby permitting full 4D control over network vascularity, potentially even in the presence of live cells. Despite this unmatched potential, the complete benefit of using light to generate vascular networks within tissue-engineered constructs has not yet been fully realized. Subtractive strategies employing light to date have relied on photoablative methods, whereby large intensities of high-energy light induces material degradation through non-specific chemical bond photolysis, extreme local heating, or microcavitation.[18,19] Though effective in controlling 3D channel geometry, the use of photoablation raises serious concerns over the effects of ablative laser light on cellular integrity and processes while opening unanswered questions regarding the integrity of surrounding materials. To truly exploit the potential of light-based subtraction for vascular engineering, strategies that employ cytocompatible wavelengths and intensities of light to create multicellular 3D tissues remain in great need.
Herein we introduce a fully cytocompatible fabrication strategy involving precise molecular photolysis to generate complex 3D vascular networks within hydrogel biomaterials. Demonstrated in this work is a versatile class of synthetic peptide-polymer-based materials that supports cellular encapsulation, enzyme-mediated matrix remodeling, and biochemical customization with the capacity for photoprogrammable vascular formation. Through multiphoton lithography-assisted photoscission, perfusable vessels are created within photosensitive gels with features that span nearly all physiological size ranges of interest, hierarchical organization, and complete 4D control. It is further shown that photodegraded channels can be readily modified over time and to display user-defined intraluminal topographies, providing a new and unique route to regulate local cell functions through dynamic and specific biophysical interactions. Finally, we exploit these customizable materials to fabricate the smallest endothelialized vessels to date as well as multilayered vascular structures of unprecedented complexity in the presence of encapsulated stromal cells, signifying crucial steps towards engineering of functional synthetic tissue.
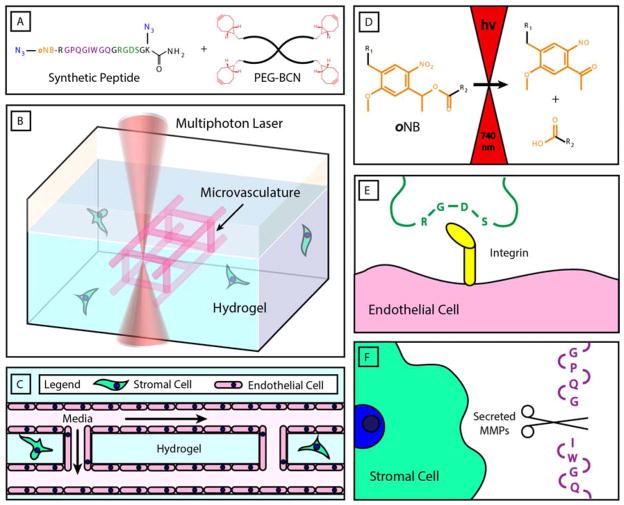
Photodegradable hydrogels were generated by strain-promoted azide-alkyne cycloaddition (SPAAC) between poly(ethylene glycol) tetrabicyclononyne (PEG-tetraBCN, Mn ~ 20,000 Da) and a diazide-functionalized synthetic peptide [N3-oNB-RGPQGIWGQGRGDSGK(N3)-NH2] (Figure 1A). The bioorthogonal nature of the SPAAC reaction enables cells to be encapsulated during hydrogel synthesis with high viability (Figure 1B–C).[17,20,21] SPAAC-based networks form rapidly via a step-growth polymerization mechanism, ensuring that all material crosslinks include functionalities introduced by the synthetic peptide. Critically, the peptide crosslinker contains a photodegradable ortho-nitrobenzyl ester (oNB) moiety that undergoes defined molecular photolysis in response to pulsed near infrared light (λ = 740 nm), enabling programmable vessel formation using cytocompatible light within the optical window of biological tissue (Figure 1D).[16,17,21,22] The peptide is also engineered to contain the fibronectin-derived, integrin-binding RGDS peptide sequence (italicized),[23] known to promote general cell attachment, spreading, and proliferation (Figure 1E). RGDS is of particular interest in vascular engineering as it is binds the highly expressed and angiogenesis-mediating αVβ5 and αVβ3 integrins, critically associated with capillary sprouting and vascular endothelial growth factor receptor-2 regulation.[24,25] Finally, the crosslinker contains the GPQGIWGQ polypeptide sequence (underlined) that is susceptible to enzymatic cleavage by a wide variety of matrix metalloproteinases (MMPs),[26] ultimately enabling cell-mediated gel remodeling (Figure 1F). These combined aspects provide for a material that is cytocompatible, promotes endothelialization, supports matrix remodeling by encapsulated cells, and enables user-programmable vessel formation through well-defined photodegradation chemistry. Though photodegradable hydrogels have demonstrated substantial utility in dynamic cell culture,[16,17,27,28] their usage has been confined to bulk material softening, surface erosion, or void creation isolated to locations within materials; perfusable vessel structures have not yet been created via biomaterial photodegradation.
Figure 1.
Microvasculature fabrication within multifunctional hydrogel biomaterials. A) Hydrogels are generated by cytocompatible strain-promoted azide-alkyne cycloaddition (SPAAC) between a poly(ethylene gycol) tetrabicyclononyne (PEG-tetraBCN, where BCN is shown in red) and a diazide-modified synthetic peptide (azides shown in blue). The peptide contains a photodegradable ortho-nitrobenzyl linker (oNB, orange), a matrix metalloproteinase (MMP)-cleavable sequence (GPQGIWGQ, purple), and a cell-adhesive region (RGDS, green). B) User-programmed multiphoton excitation induces localized degradation of the hydrogel through oNB photocleavage, resulting in the microchannel (pink) generation in the presence of encapsulated stromal cells (green). C) Photodegraded networks are then seeded with endothelial cells (pink) to created cell-laden hydrogels with perfusable, endothelialized vasculature. D) oNB photocleavage yields nitroso- and acid-terminated byproducts. E) RGDS moieties included within the peptide crosslinker promote cell attachment and proliferation. F) The GPQGIWGQ peptide sequence is cleaved enzymatically by secreted MMPs, enable cell-mediated matrix remodeling of the hydrogel network.
To control gel photodegradation and vessel formation over single micron-resolved volumes while maximizing cytocompatibility, multiphoton lithography was employed. As multiphoton excitation requires near-simultaneous absorption of two independent photons, photochemically initiated events are largely confined to the laser focal point, thereby affording sub-micron control in all three spatial dimensions. Furthermore, by employing pulsed near infrared light whose wavelength falls within the optical window of biological tissue, precise photochemistries can be performed deep within materials while minimizing phototoxicity concerns for cells above and below the focal point. Since the photolabile oNB moiety is included in every hydrogel crosslink, complete photodegradation occurs rapidly as guided by programmed laser rastering and a computer-controlled piezoelectric stage. Though the theoretical x- and y-patterning resolution of this strategy is limited only by the wavelength of light employed, z-resolution and fabrication depth limitations are governed practically by the utilized optics (e.g., working distance of objective, laser power, wavelength). To permit real-time visualization of material photodegradation and channel formation, gels were fluorescently labeled with an azide-functionalized fluorescein during formulation. Channel patency and perfusability were then confirmed post-fabrication by flowing and nonspecifically coating channel walls with red-fluorescent polystyrene beads (FluoSpheres®, 0.22 μm or 2 μm diameter, Invitrogen) or direct endothelial seeding within channel lumens.
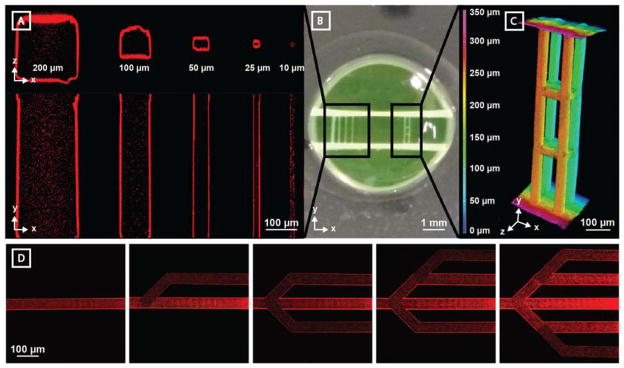
In order to demonstrate the wide range over which multiphoton photolysis can be utilized to create perfusable channels, five photodegraded vessels of decreasing widths and heights were generated in the Y-direction between two large inlet/outlet channels (500 μm diameter, separated by ~1.5 mm) introduced by needle-based subtraction molding in the X-direction (Figure 2A–B). Microvessels with cross sections as large as 200 μm x 200 μm and as small as 10 μm x 10 μm were generated. Fluorescent imaging of the entire channel lengths revealed complete material degradation, removal of sol material for all sizes, and facile perfusion with 0.22 μm diameter fluorescent beads. Perfusable channels could be photogenerated at any z-location throughout the entire gel thickness (1.2 mm, Supplementary Figure S1) without altering photodegradation parameters (e.g., laser intensity, voxel dwell time). To demonstrate that similar control over vessel structure is afforded in 3D, four parallel channels were generated in the Y-direction between the inlet and outlet channels, and then joined together at two locations by four interconnecting channels in the X-direction, and four interconnecting channels in the Z-direction (all with 50 μm x 50 μm cross sections, Figure 2B–C). Upon perfusion with fluorescent beads, intact lumens were observed with clear interconnecting channels in all three X, Y, and Z planes. As photodegradation can be initiated on demand, we sought to demonstrate that network structure could be evolved over time post-fabrication. Interconnected channels (40 μm x 40 μm cross sections) were modified sequentially through successive material photodegradation (Figure 2D). Channel patency was confirmed by fluorescent bead perfusion following iterative network alteration. Collectively, these studies highlight the ability to create and modify perfusable synthetic vasculature of user-defined size and geometry with unmatched 4D control.
Figure 2.
Microvessel generation is readily controlled in 3D with micron-scale resolution. Vertical channel sets were generated between two horizontal channels (~500 μm diameter) spaced ~1.5 mm apart. Vessel perfusion with fluorescent polymer beads (red, 0.22 μm diameter) enabled channel visualization by multiphoton microscopy. A) Channels with a wide range of cross-sectional sizes (200 μm x 200 μm, 100 μm x 100 μm, 50 μm x 50 μm, 25 μm x 25 μm, and 10 μm x 10 μm) are fully patent. B) Photographic image of hydrogel displays both parallel microchannel and 3D multilayered channel sets. C) Photodegraded channels can be generated with full 3D control. Four parallel 50 μm x 50 μm (width x height) channels generated in the Y direction (X-spacing = 150 μm, Z-spacing = 100 μm) connect the horizontal channels. 8 bridging channels (50 μm x 50 μm, cross sectional) interconnect features in the X and Z directions. Microbead fluorescence is viewed using Z-depth color-coding. D) Perfusable vascular network geometry can be altered over time through iterative network photodegradation (channel cross section of 40 μm x 40 μm, red beads = 0.22 μm diameter).
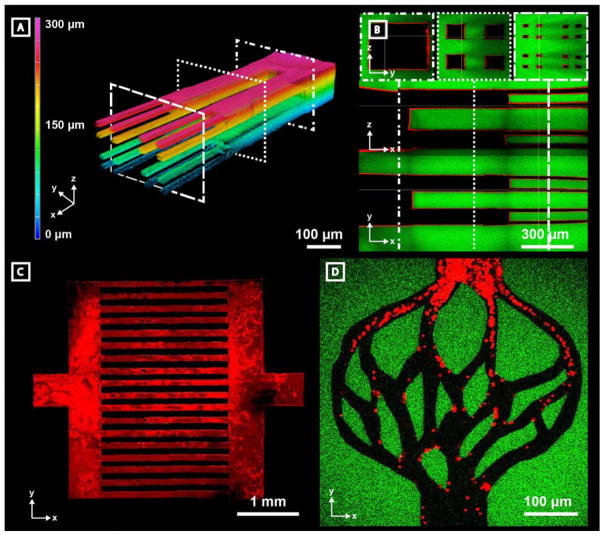
Critical to the development of large-scale engineered tissue is the capacity to support cell metabolism through strategies extending past simple diffusion. In normal physiology, this is accomplished by a complex structural hierarchy of blood vessels where 3D branching allows for size-dependent function and directional flow control. Conversely, disturbance of physiological flow patterns as a consequence of pathologies like tumor growth can lead to abnormal neovascularization, disruption of endothelial function, and redistribution of blood flow.[29,30] Though recapitulation of vascular geometry has proven difficult to achieve through conventional strategies (e.g., soft lithography, additive manufacturing), vessel customization afforded through channel photodegradation opens new doors towards recreating physiologically relevant structures. To demonstrate the unique capacity to create complex hierarchical vessel networks in a controlled 3D manner, a single channel (300 μm x 300 μm cross section) was generated in series with four smaller channels (100 μm x 100 μm cross sections) connected to sixteen even smaller channels (25 μm x 25 μm cross sections) (Figure 3A–B). This vascular network captures the structural complexity of blood vessel hierarchy, allows for study of complex 3D flow, and accentuates the power of multiphoton degradation in vascular construction and design.
Figure 3.
Microvessels generated by programmable photodegradation exhibit complex hierarchy, scalability, and biomimetic design. Channels were generated in fluorescein-modified hydrogels (green) and perfused with fluorescent microbeads (red, 0.22 μm diameter in A–C, 2 μm in D). A) A 3D hierarchical vascular network is visualized using Z-depth color-coding of fluorescent beads. Device is comprised of one 300 μm x 300 μm channel which branches into four 100 μm x 100 μm channels and then terminates in 16 channels, each with 25 μm x 25 μm cross section. Each segment was approximately 500 μm in length in the X dimension. Corresponding dashed squares indicate respective cross sections demonstrated in panel B. B) Cross-sectional views of the branched microvascular network indicate intact vascular lumens. C) A centimeter-sized device was generated by creating two 500 μm x 500 μm x 100 μm inlets and outlets connected by two 500 μm x 3.5 mm x 100 μm distributing channels joined by 18 individual 2.5 mm x 100 μm x 100 μm (X x Y x Z dimensions) connecting channels. D) Biomimetic vascular networks can be created with user-defined geometrical control.
In addition to creating hierarchical 3D structure, successful generation of vascular constructs for tissue engineering applications requires process scalability. To assess whether such scalability is afforded by photodegradation-based vessel fabrication, rapid prototyping of cm-sized fluidic networks was attempted. Figure 3C illustrates a perfusable device consisting of a single inlet and outlet (500 μm x 100 μm cross sections) connected by a distributing channel (500 μm x 3.5 mm x 100 μm) in series with eighteen additional smaller channels (2.5 mm x 100 μm x 100 μm). Furthermore, as overall channel geometry and architecture is fully user defined by controlled laser scanning, it is demonstrated that one can readily form biomimetic vascular networks that recapitulate native vasculature (Figure 3D). After biomimetic construct formation, red fluorescent beads (2 μm diameter) roughly the size scale of platelets were perfused and imaged as they travelled through the vascular network. Though material processing is likely to be slower than other techniques to generate large-scale vascular networks (minutes to hours depending on network design and light parameters), overall process cytocompatibility enables large networks to be generated without uncontrolled cellular perturbation. Together, these data demonstrate that multiphoton photodegradation enables stable vasculature to be generated that recapitulates both the physiological size and complexity of native blood vessel networks.
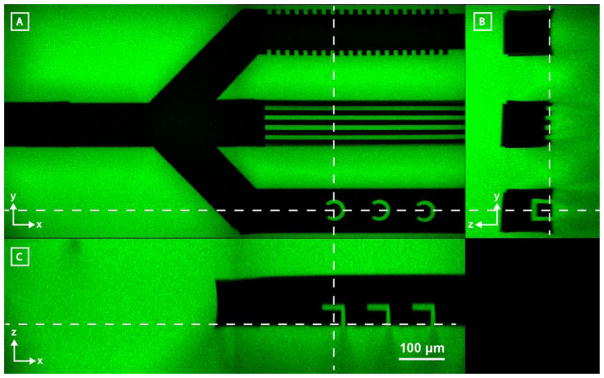
While other techniques have demonstrated the ability to control channel network design, photodegradation-based subtractive strategies further and uniquely enable micromachining of intraluminal topography. Inspired by recent findings that topographical cues and geometries can alter cell adhesion, migration, differentiation, cytoskeletal organization, and morphology,[31,32] we sought to embed such physical cues locally within devices. Further recognizing a growing interest in creating cell capture-devices[33,34] to examine single cell function and behavior, we also sought to engineer customizable microtraps within channels. Though both tasks have proven impossible using conventional techniques, we hypothesized that such architectures could be generated readily within the lumen of microfluidic channels by multiphoton photolithography. In Figure 4, a microfluidic network consisting of a single inlet and three outlets was generated within a fluorescent hydrogel. Different intravessel architectures were sculpted into each of the three outlets. The first two outlets were created with parallel microgrooves (10 μm width) perpendicularly oriented between individual outlets, highlighting the ability to pattern channel microtopography. Within the third outlet, intravessel cell traps (~50 μm size scale) were successfully created and used for cell isolation (Supplementary Figure S2), demonstrating the capacity to micromachine 3D functional capture devices. We anticipate that such topographical microarchitectures will provide vascular engineers with new tools to simultaneously control endothelial cell function and study vessel structure, while probing perfusate and blood constituents in a novel and user-designed manner.
Figure 4.
Photodegradation-mediated microfabrication of intraluminal channel architectures within hydrogel biomaterials (green). A–C) Z, Y, X cross-sectional views of a three-branched channel modified with different intraluminal architectures. The top channel has been micromachined to contain microgrooves in the XZ plane, the middle microgrooves in the XY plane, and the bottom cylindrical cell traps. Dashed lines indicate cross-sectional areas as observed in each of the three perspective views. All images are to scale.
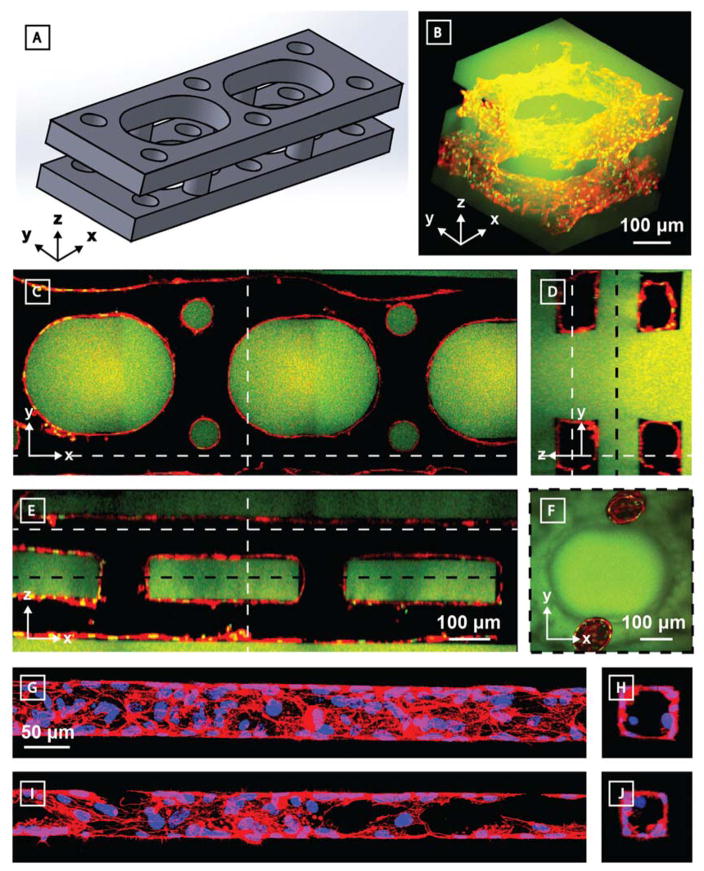
Native vascular networks are not only complex and hierarchical, but also endothelial-lined. To demonstrate that our photodegradable material is capable of generating and supporting 3D microvasculature, human umbilical vein endothelial cells (HUVECs) were perfusion seeded within the lumen of photodegraded channels generated in fluorescein-modified PEG hydrogels (Figure 5). Within these devices, two identical layers of parallel vessels (~100 μm x 100 μm cross sections) were stacked directly above each other (100 μm separation in the Z direction). Each layer was constructed with curved interconnecting channels in the XY plane (Figure 5A–E) while the two layers were connected together by circular vertical channels in the Z direction (100 μm in diameter) (Figure 5F). Curved interconnected channels were selected to encourage overall fluid flow within the device, increasing efficiency of cell seeding and perfusion through channels while limiting cellular accumulation and potential channel clogging, as well as to promote overall endothelial attachment and function.[35] Ten days following endothelialization, samples were fixed, stained for cytoskeletal actin (Alexa Fluor 532 Phalloidin) and nuclei (Hoescht 33342), and imaged by multiphoton fluorescent microscopy. Seeded HUVECs attached and spread to cover the walls of the photodegraded channel with well-formed lumens, indicating successful generation of endothelialized vessels. Finally, to probe the spatial limitations of endothelialization, microchannels with even smaller features sizes (cross sections of 45 μm x 45 μm, 60 μm x 60 μm) were also demonstrated (Figure 5G–J, Supplementary Figure S3). To our knowledge, this represents the smallest endothelialized synthetic vessel to date within a material that supports cell encapsulation.
Figure 5.
3D endothelialized channels generated within photodegradable fluorescent gels (green). Two identical parallel-oriented channels are interconnected with cylindrical channels in the Y-direction. 10 days following microvessel endothelialization with HUVECs, samples were fixed and stained prior to imaging by multiphoton microscopy. F-actin is shown in red, nuclei in green. A) 3D model of patterned channels. Endothelialized sample is visualized B) as a 3D render and C–F) as Z-, X-, and Y-cross sectional views. Dashed white lines indicate cross-sectional areas observed. An additional Z-cross-sectional area between the two channel sets (location denoted by dashed black line) demonstrates interconnectivity in the Z direction. Endothelialization of G–H) 60 μm x 60 μm and I–J) 45 μm x 45 μm (width x height) channels was obtained. F-actin is shown in red, nuclei in blue. G) and I) represent XY projections; H) and J) are YZ slices. C–E) are all to scale, as are G–J).
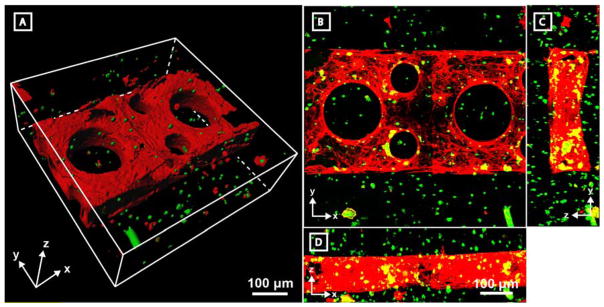
Though synthetic vascular beds are extremely valuable tools in vascular biology and hemodynamics, the most basic and critical roles of blood vessels is to support complex organ function. To date, the ability to create 3D multicellular vascularized tissues remains one of the most critical – if not the most critical – engineering feats hindering the use of metabolically demanding engineered tissues clinically. While there are few demonstrations of controlled 3D endothelialization, there are fewer still that permit simultaneous encapsulation of cells in the stroma and endothelialization without incurring cellular damage. To highlight that photodegradation can be performed in a cytocompatible manner, a single layer set of parallel channels with rounded interconnecting channels was generated in the presence of human bone marrow-derived hS5 stromal cells constitutively expressing green fluorescent protein (GFP). Four days after endothelialization with HUVECs, samples were fixed and subsequently imaged (Figure 6). hS5 cells above and below the photodegraded channels remained viable during the entire culture time course, as evidenced by their continued production of GFP as well as quantitative LIVE/DEAD analysis (Supplementary Figure S4). The ability to create customizable endothelialized vessels in the presence of encapsulated cells represents a powerful step towards engineering complex heterogeneous tissue for clinical transplantation. Ongoing work seeks to explore both the enzyme-mediated matrix remodeling of encapsulated cells and endothelial cells of differing origin in these matrices. Furthermore, interest exists in tailoring matrix sensitivity to endothelial cell specific MMP secretion.
Figure 6. Endothelialized channels are readily fabricated in the presence of encapsulated stromal cells.
A single-layer channel was generated by photodegradation in the presence of encapsulated GFP-expressing hS5 stromal cells (green). Channels were then endothelialized with HUVECs, cultured for 4 days, fixed, and stained for F-actin (red) prior to imaging by fluorescent multiphoton microscopy. Co-stained and single-stained green cells are identified as hS5 cells while singly red-stained cells represent HUVECs. Sample is viewed A) as a 3D surface render of a 300 μm Z-stack and B–D) as Z-, X-, and Y-direction maximum intensity projections. B–D) are all to scale.
As demonstrated, multiphoton photodegradation revels in its ability to generate high-resolution vascular features with 4D structural control in a cytocompatible and programmable manner. While this strategy does grant users the ability to both prototype vessel designs and explore geometries that have been previously unattainable, it is by nature a low-throughput fabrication technique that is not without its own limitations. Though large vessels can be generated by controlled photolysis, because the process is one of material subtraction, fabrication time scales with vessel size; increased processing times practically limits vessel diameters to the sub-mm range. In cases where both large and small vessels are of interest, multiscale vessel creation involving complementary fabrication strategies performed in tandem may prove useful; higher-throughput techniques (e.g., soft lithography) can be used to create large vessels within photodegradable materials which can be subsequently outfitted with photopatterned microchannels. Furthermore, because molecular photoscission is aided through a repurposed multiphoton microscope, certain restrictions are imposed. Depth of penetration into samples, fabrication speed, and efficiency of photodegradation are limited by the working distance, spot size, and numerical aperture of the objective lenses utilized, as well as the power of the laser source employed. Further customization of both the equipment and software could enhance vessel generation capacity. Though photodegradation-mediated vessel fabrication is not without its own limitations, abundant opportunities to mitigate these challenges exist.
As previously outlined, the development of biomimetic, multicellular, and clinically relevant engineered tissues requires scalable methodology to generate 3D vascular beds. Such methods must be cytocompatible, enable creation of vessels that span the geometries of all native vasculature, and permit controlled biochemical and biophysical interactions between the biomaterial and endothelium. The novel photodegradable polymer-peptide hydrogel that we have presented here satisfies each of these requirements with exquisite synthetic and spatial control. Multiphoton-assisted photodegradation enables fabrication and subsequent modification of complex endothelialized 3D microvascular networks with customizable intraluminal architectures in the presence of encapsulated cells. We expect such biomimetic hierarchical vascular networks to provide new opportunities towards generation of complex multicellular tissues with defined architectures and physiologically relevant thicknesses, as well as for investigating the effects of vascular flow on endothelialization, studying blood-capillary interactions, and recapitulating dense vascular networks to support metabolically demanding cells.
Experimental Section
Synthesis of Diazide Peptide Crosslinker
H-RGPQGIWGQGRGDSGK(N3)-NH2 peptide was synthesized (0.5 mmol scale) by standard automated microwave-assisted Fmoc solid-phase methodologies (CEM Liberty 1). Synthesis was performed on Rink amide resin (ChemPep) using a C-terminal Fmoc-Lys(N3)-OH (synthesized via known synthetic routes[36,37]). N-terminal couplings were performed by treating resin with protected amino acid (2 mmol), 2-(1H-benzotriazol-1-y1)-1,1,3,3-tetramethyluronium hexafluorophosphate (2 mmol), and N,N-diisopropylethylamine (2 mmol, DIEA) in a solution of dimethylformamide (DMF) and N-Methyl-2-pyrrolidone (9 mL and 2 mL, respectively) at 75 °C for 5 min. Fmoc deprotection reactions were performed using 1-hydroxybenzotriazole (0.1 M), piperidine in DMF (20 v/v%) at 90 °C for 90 s. Photodegradable azide functionality was then conjugated to the peptide’s N-terminal amine by treating resin overnight with N3-oNB-OSu[17] (0.65 mmol, 330 mg) and DIEA (2 mmol, 258 mg) in minimal DMF. Peptide deprotection and cleavage from resin was performed by two-hour treatment in trifluoroacetic acid (TFA)/triisopropylsilane/water (95:2.5:2.5 v/v%, 30 mL). Crude product was precipitated in and washed twice with ice-cold diethyl ether. The crude peptide was then purified using preparatory high performance liquid chromatography (HPLC, C18 column) utilizing a 43.4 minute linear gradient (20–100%) of acetonitrile in aqueous TFA (0.1 v/v%). The final product was then lyophilized to yield a dark-yellow solid, denoted as N3-oNB-RGPQGIWGQGRGDSGK(N3)-NH2. Peptide purity was confirmed by MALDI-TOF: Calculated for C86H129N32O29+ [M + 1H]+, to be 2073.9; observed 2072.6.
Hydrogel Formation
Hydrogels were formed by combining PEG-tetraBCN (Mn ~ 20,000 Da, synthesized as previously described)[21] with the diazide N3-oNB-RGPQGIWGQGRGDSGK(N3)-NH2 peptide in a 1:2 stoichiometric ratio in either phosphate buffered saline (PBS, pH = 7.4) or stromal cell culture media (described below) to achieve a final 7 wt% of prepolymer solution. This mixture was then quickly pipetted between two glass coverslips (one Rain-X® treated, the other untreated) separated by silicone spacers (thickness = 500 μm) and allowed to react for 1 hour at room temperature. After gelation, the Rain-X® coated coverslip was removed, yielding hydrogels of defined thickness that remained bound to the untreated glass coverslip. For fluorescein-conjugated hydrogels, PEG-tetraBCN (0.01 mmol) was pre-reacted with 5-FAM azide (AAT Bioquest, 0.002 mmol) in PBS for 2 hours at room temperature. Product was then dialyzed (MWCO ~ 1kDa, Spectra/Por® 7) against water, lyophilized, and reconstituted in PBS. To form fluorescent hydrogels, FAM-conjugated PEG-tetraBCN was mixed with unconjugated PEG-tetraBCN in a 1:10 ratio and then reacted with diazide peptide crosslinkers as described above.
Multiphoton Photodegradation
Photodegradation was performed by multiphoton lithography using an Olympus FV1000 MPE BX61 Microscope equipped with a variable wavelength Mai Tai DeepSee Ti:S laser with an maximum power of 2.7W and a water-immersion objective lens (25x, NA = 1.05) (Garvey Imaging Center, University of Washington) in conjunction with Olympus FluoView imaging software. Channel patterns within a single field of view (500 μm x 500 μm) were generated by defining regions of interest followed by laser rastering to intended Z-depths (Z-step size = 1 μm). Networks larger than a single field of view were programmed by stitching together single fields of view with defined regions of interest using the FluoView mosaic feature. Individual scans were performed at λ = 740 nm using 1.9W laser power with a pixel dwell time of 2 μs and with 5–7 frame repeat scans to ensure complete photodegradation.
Endothelialization of Photodegraded Channels
HUVECs were cultured in Endothelial Cell Basal Media (Lonza) supplemented with Endothelial Cell Growth Media (EGM) SingleQuots (Lonza). Cell cultures were passaged upon reaching 80% confluency onto gelatin-coated tissue culture flasks and were used prior to passage 8. Seeding of endothelial cells within channel lumens was performed by trypsin treatment (0.05 mM in PBS, 5 min) of HUVEC monolayers, centrifugation, and media washing, followed by direct pipetting of cell solution (5 x 107 cells mL−1) onto device microchannel inlets. Capillary action and differential pressure pushed the cell-rich solution through the channel network. Cells were allowed to attach to vessel walls for 1 hour, rinsed twice with growth media, and maintained in culture (37 °C, 5% CO2).
3D Cell Encapsulation and Cell Culture
Human bone marrow-derived hS5 stromal cells constitutively expressing GFP were cultured in RPMI 1640 basal media (Corning) supplemented with fetal bovine serum (10%, Corning) and penicillin/streptomycin (1%, Corning). Cells were trypsinized and passaged upon reaching 80% confluency. To prepare cells for encapsulation, stromal cells were treated with trypsin (0.05 mM in PBS, 5 min), centrifuged (1,200 RPM for 4 mins), and resuspended in hydrogel precursor formulations (5 x 106 cells mL−1). Mixtures were gently vortexed to ensure homogenous encapsulation, pipetted onto coverslips, and allowed to gel at room temperature (1 hr). After microchannel fabrication and endothelialization, hydrogels containing encapsulated hS5 stromal cells and HUVEC endothelialized vessels were maintained using EGM SingleQuot-supplemented Endothelial Growth Basal Media.
Cellular Staining and Visualization
Constructs were fixed by replacing culture media with and gently perfusing formaldehyde in PBS (3.7 wt/vol %, 30 min). Fixative was removed by performing three PBS washes (5 min each). To minimize nonspecific binding and to permeabilize cells, constructs were incubated in PBS containing bovine serum albumin (BSA, 3 wt/vol %) and Triton X-100 (1 vol/vol %) for 1 hr. Samples were then stained using either or both Alexa Fluor 532 Phalloidin (1:100, Thermo Fisher Scientific) and Hoescht 33342 (1:1000, Thermo Fisher Scientific) in PBS supplemented with BSA (1 wt/vol %). Imaging was performed by multiphoton microscopy (Olympus FV1000 MPE BX61), with data analysis and visualization performed with FluoView (Olympus), ImageJ (NIH), and Imaris software (Bitplane).
Supplementary Material
Figure S1. Photodegraded microvasculature can be generated throughout thick samples. Channels (30 μm x 30 μm cross sections) were generated in PEG-peptide hydrogels at increments of 100 μm in the Z direction and then filled with red fluorescent beads (0.22 μm diameter). Vascularized constructs were imaged by multiphoton microscopy and displayed as a maximum intensity projection across the XZ plane. Perfusable channels were observed throughout the entire thickness of the sample.
Figure S2. Intraluminal single cell traps. Photodegraded channels (300 μm x 100 μm cross section) were generated containing single-cell capture devices (outer diameter = 100 μm, inner diameter = 50 μm) A) without or B) with an outlet to encourage flow through the capture device. Perfusion of cell suspensions (106 hS5 cells mL−1) in stromal cell growth media revealed individual cell capture events, as imaged by brightfield microscopy.
Figure S3. Sub-100 μm photodegradated channels are readily endothelialized. A) 60, B) 45, and C) 30 μm square cross-sectional photodegraded channels were generated in non-fluorescent hydrogels and subsequently endothelialized. Brightfield Images were acquired 6 hours post endothelialization. All images are to scale.
Figure S4. Encapsulated stromal cells remain viable above and below areas of microchannel fabrication. Channel fabrication was performed in cell-laden hydrogels containing encapsulated hS5 stromal cells (5 x 106 cells mL−1) and then stained with a LIVE/DEAD kit (Invitrogen, green = live, red = dead). No significant difference in viability was observed for regions above vessel patterning compared with those elsewhere in the material.
Movie S1. Endothelialized channels are readily fabricated in the presence of encapsulated stromal cells.
Rotating 3D view of endothelialized channels fabricated in the presence of encapsulated stromal cells. Data was visualized using the “Surface” function in Imaris (Bitplane). F-actin is shown in red, stromal cells in green.
Acknowledgments
The authors recognize and thank Drs. Ron Seifert and Dale Hailey of the University of Washington Garvey Imaging Center for their ongoing support and advice, as well as Dr. Brian Hayes for gifting the hS5 cells utilized in the encapsulation experiments. This work was supported by a University of Washington Faculty Startup Grant to Dr. Cole A. DeForest, a University of Washington Bioengineering Cardiovascular Training Grant (Ruth L. Kirschenstein NRSA Training Grant, T32EB001650) awarded to Mr. Christopher K. Arakawa, and grants from the National Institute of Health awarded to Mr. Christopher K. Arakawa (1F30HL134298) and to Dr. Ying Zheng (DP2DK102258).
Footnotes
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Christopher K. Arakawa, Department of Bioengineering, University of Washington, 3720 15th Ave NE, Seattle, WA 98105
Barry A. Badeau, Department of Chemical Engineering, University of Washington, 3781 Okanogan Lane NE, Seattle, WA 98195
Prof. Ying Zheng, Department of Bioengineering, University of Washington, 3720 15th Ave NE, Seattle, WA 98105. Institute for Stem Cell & Regenerative Medicine, University of Washington, 850 Republican Street, Seattle, WA 98109
Prof. Cole A. DeForest, Department of Bioengineering, University of Washington, 3720 15th Ave NE, Seattle, WA 98105. Department of Chemical Engineering, University of Washington, 3781 Okanogan Lane NE, Seattle, WA 98195. Institute for Stem Cell & Regenerative Medicine, University of Washington, 850 Republican Street, Seattle, WA 98109
References
- 1.Carmeliet P. Nat Med. 2000;6:389. doi: 10.1038/74651. [DOI] [PubMed] [Google Scholar]
- 2.Coultas L, Chawengsaksophak K, Rossant J. Nature. 2005;438:937. doi: 10.1038/nature04479. [DOI] [PubMed] [Google Scholar]
- 3.Jain RK. Nat Med. 2003;9:685. doi: 10.1038/nm0603-685. [DOI] [PubMed] [Google Scholar]
- 4.Michiels C. J Cell Physiol. 2003;196:430. doi: 10.1002/jcp.10333. [DOI] [PubMed] [Google Scholar]
- 5.Hasan A, Paul A, Vrana NE, Zhao X, Memic A, Hwang YS, Dokmeci MR, Khademhosseini A. Biomaterials. 2014;35:7308. doi: 10.1016/j.biomaterials.2014.04.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morgan JP, Delnero PF, Zheng Y, Verbridge SS, Chen J, Craven M, Choi NW, Diaz-Santana A, Kermani P, Hempstead B, López JA, Corso TN, Fischbach C, Stroock AD. Nat Protoc. 2013;8:1820. doi: 10.1038/nprot.2013.110. [DOI] [PubMed] [Google Scholar]
- 7.Jia W, Gungor-Ozkerim PS, Zhang YS, Yue K, Zhu K, Liu W, Pi Q, Byambaa B, Dokmeci MR, Shin SR, Khademhosseini A. Biomaterials. 2016;106:58. doi: 10.1016/j.biomaterials.2016.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khalil S, Sun W. J Biomech Eng. 2009;131:111002. doi: 10.1115/1.3128729. [DOI] [PubMed] [Google Scholar]
- 9.Zhu W, Qu X, Zhu J, Ma X, Patel S, Liu J, Wang P, Lai CSE, Gou M, Xu Y, Zhang K, Chen S. Biomaterials. 2017;124:106. doi: 10.1016/j.biomaterials.2017.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng Y, Chen J, Craven M, Choi NW, Totorica S, Diaz-Santana A, Kermani P, Hempstead B, Fischbach-Teschl C, López JA, Stroock AD. Proc Natl Acad Sci U S A. 2012;109:9342. doi: 10.1073/pnas.1201240109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng Y, Chen J, López JA, Paulinska P, Spiel A, Jilma B, Tsai HM, Tsai AW, De Meyer SF, De Maeyer B, Deckmyn H, Vanhoorelbeke K, Giblin JP, Hewlett LJ, Hannah MJ, Van Mourik JA, de WTR, Voorberg J, Dong JF, Di Stasio E, De Cristofaro R, Nolasco LH, Schwameis M, Schorgenhofer C, Assinger A, Steiner M, Jilma B, Chen J, Shankaran H, Alexandridis P, Neelamegham S, Zheng Y, Han HC, Liu Q, Mirc D, Fu BM, Guglielmini L, Rusconi R, Lecuyer S, Stone HA, Furlan M, Robles R, Lamie B, Shim K, Anderson PJ, Tuley EA, Wiswall E, Sadler JE, Levy GG, Kokame K, Furlan M, Tsai HM, Lian EC, Chung DW, Uemura M, McCarty OJ, Schouten M, Wiersinga WJ, Levi M, van der Poll T, Grau GE, Wolinsky H, Cross VL. Nat Commun. 2015;6:7858. [Google Scholar]
- 12.Lee VK, Kim DY, Ngo H, Lee Y, Seo L, Yoo SS, Vincent PA, Dai G. Biomaterials. 2014;35:8092. doi: 10.1016/j.biomaterials.2014.05.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bersini S, Yazdi IK, Talò G, Shin SR, Moretti M, Khademhosseini A. Biotechnol Adv. 2016;34:1113. doi: 10.1016/j.biotechadv.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen DHT, Cohen DM, Toro E, Chen AA, Galie PA, Yu X, Chaturvedi R, Bhatia SN, Chen CS. Nat Mater. 2012;11:768. doi: 10.1038/nmat3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nahmias Y, Schwartz RE, Verfaillie CM, Odde DJ. Biotechnol Bioeng. 2005;92:129. doi: 10.1002/bit.20585. [DOI] [PubMed] [Google Scholar]
- 16.Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Science. 2009;324:59. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeForest CA, Anseth KS. Nat Chem. 2011;3:925. doi: 10.1038/nchem.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brandenberg N, Lutolf MP. Adv Mater. 2016;28:7450. doi: 10.1002/adma.201601099. [DOI] [PubMed] [Google Scholar]
- 19.Heintz KA, Bregenzer ME, Mantle JL, Lee KH, West JL, Slater JH. Adv Healthc Mater. 2016;5:2153. doi: 10.1002/adhm.201600351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeForest CA, Polizzotti BD, Anseth KS. Nat Mater. 2009;8:659. doi: 10.1038/nmat2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeForest CA, Tirrell DA. Nat Mater. 2015;14:523. doi: 10.1038/nmat4219. [DOI] [PubMed] [Google Scholar]
- 22.DeForest CA, Anseth KS. Angew Chemie - Int Ed. 2012;51:1816. doi: 10.1002/anie.201106463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joshi P, Chung CY, Aukhil I, Erickson HP. J Cell Sci. 1993;389 doi: 10.1242/jcs.106.1.389. [DOI] [PubMed] [Google Scholar]
- 24.Iivanainen E, Kähäri V-M, Heino J, Elenius K. Microsc Res Tech. 2003;60:13. doi: 10.1002/jemt.10238. [DOI] [PubMed] [Google Scholar]
- 25.Somanath PR, Malinin NL, Byzova TV. Angiogenesis. 2009;12:177. doi: 10.1007/s10456-009-9141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagase H, Fields GB. Biopolymers. 1996;40:399. doi: 10.1002/(SICI)1097-0282(1996)40:4%3C399::AID-BIP5%3E3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 27.Yang C, Tibbitt MW, Basta L, Anseth KS. Nat Mater. 2014;13:645. doi: 10.1038/nmat3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tibbitt MW, Kloxin AM, Dyamenahalli KU, Anseth KS. Soft Matter. 2010;6:5100. doi: 10.1039/C0SM00174K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jain RK, Stylianopoulos T. Nat Rev Clin Oncol. 2010;7:653. doi: 10.1038/nrclinonc.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jain RK. Science. 2005;307:58. doi: 10.1126/science.1104819. [DOI] [PubMed] [Google Scholar]
- 31.Kim DH, Han K, Gupta K, Kwon KW, Suh KY, Levchenko A. Biomaterials. 2009;30:5433. doi: 10.1016/j.biomaterials.2009.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim DH, Lipke EA, Kim P, Cheong R, Thompson S, Delannoy M, Suh KY, Tung L, Levchenko A. Proc Natl Acad Sci U S A. 2010;107:565. doi: 10.1073/pnas.0906504107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin CH, Hsiao YH, Chang HC, Yeh CF, He CK, Salm EM, Chen C, Chiu IM, Hsu CH. Lab Chip. 2015;15:2928. doi: 10.1039/c5lc00541h. [DOI] [PubMed] [Google Scholar]
- 34.Kim HS, Devarenne TP, Han A. Lab Chip. 2015;15:2467. doi: 10.1039/c4lc01316f. [DOI] [PubMed] [Google Scholar]
- 35.Esch MB, Post DJ, Shuler ML, Stokol T. Tissue Eng Part A. 2011;17:2965. doi: 10.1089/ten.tea.2010.0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goddard-Borger ED, Stick RV. Org Lett. 2007;9:3797. doi: 10.1021/ol701581g. [DOI] [PubMed] [Google Scholar]
- 37.Chapman R, Jolliffe KA, Perrier S. Aust J Chem. 2010;63:1169. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Photodegraded microvasculature can be generated throughout thick samples. Channels (30 μm x 30 μm cross sections) were generated in PEG-peptide hydrogels at increments of 100 μm in the Z direction and then filled with red fluorescent beads (0.22 μm diameter). Vascularized constructs were imaged by multiphoton microscopy and displayed as a maximum intensity projection across the XZ plane. Perfusable channels were observed throughout the entire thickness of the sample.
Figure S2. Intraluminal single cell traps. Photodegraded channels (300 μm x 100 μm cross section) were generated containing single-cell capture devices (outer diameter = 100 μm, inner diameter = 50 μm) A) without or B) with an outlet to encourage flow through the capture device. Perfusion of cell suspensions (106 hS5 cells mL−1) in stromal cell growth media revealed individual cell capture events, as imaged by brightfield microscopy.
Figure S3. Sub-100 μm photodegradated channels are readily endothelialized. A) 60, B) 45, and C) 30 μm square cross-sectional photodegraded channels were generated in non-fluorescent hydrogels and subsequently endothelialized. Brightfield Images were acquired 6 hours post endothelialization. All images are to scale.
Figure S4. Encapsulated stromal cells remain viable above and below areas of microchannel fabrication. Channel fabrication was performed in cell-laden hydrogels containing encapsulated hS5 stromal cells (5 x 106 cells mL−1) and then stained with a LIVE/DEAD kit (Invitrogen, green = live, red = dead). No significant difference in viability was observed for regions above vessel patterning compared with those elsewhere in the material.
Movie S1. Endothelialized channels are readily fabricated in the presence of encapsulated stromal cells.
Rotating 3D view of endothelialized channels fabricated in the presence of encapsulated stromal cells. Data was visualized using the “Surface” function in Imaris (Bitplane). F-actin is shown in red, stromal cells in green.