Abstract
Background
A number of reports have described the protective effects of ginsenoside Rg1 (Rg1) in Alzheimer's disease (AD). However, the protective mechanisms of Rg1 in AD remain elusive.
Methods
To investigate the potential mechanisms of Rg1 in β-amyloid peptide-treated SH-SY5Y cells, a comparative proteomic analysis was performed using stable isotope labeling with amino acids in cell culture combined with nano-LC-MS/MS.
Results
We identified a total of 1,149 proteins in three independent experiments. Forty-nine proteins were significantly altered by Rg1 after exposure of the cells to β-amyloid peptides. The protein interaction network analysis showed that these altered proteins were clustered in ribosomal proteins, mitochondria, the actin cytoskeleton, and splicing proteins. Among these proteins, mitochondrial proteins containing HSD17B10, AARS2, TOMM40, VDAC1, COX5A, and NDUFA4 were associated with mitochondrial dysfunction in the pathogenesis of AD.
Conclusion
Our results suggest that mitochondrial proteins may be related to the protective mechanisms of Rg1 in AD.
Keywords: Alzheimer's disease, ginsenoside Rg1, mitochondria, proteomics, SILAC
1. Introduction
Alzheimer's disease (AD) is the most common neurodegenerative disorder, usually occurring in people older than 65 years [1]. AD is pathologically characterized by brain shrinkage, amyloid-rich senile plaques, and neurofibrillary tangles [2]. The clinical symptoms of AD are loss of memory, poor recognition ability, changes in personal character, and loss of body control [3], [4]. It has been reported that the main cause of AD is the accumulation of β-amyloid peptides (Aβ) [5]. The Aβ is produced from the amyloid precursor protein, which is cleaved by β-secretase and γ-secretase [6]. The cleaved Aβ gives rise to oligomeric forms, which form plaques in the brain [7]. The plaque formations trigger the pathological cascade in AD [4]. Aβ25–35, a very short fragment of Aβ, exhibits large β-sheet fibrils and retains the toxicity of the full-length peptides. Thus, Aβ25–35 represents the biologically active region of Aβ [8]. It has been reported that Aβ25–35 leads to mitochondrial dysfunctions by blocking the entrance of nuclear-encoded proteins into mitochondria, which diminishes the mitochondrial membrane potential and alters mitochondrial morphology [9].
Ginseng has been a popular and widely used traditional herbal medicine in Asia for thousands of years. Previous studies have suggested that the components of ginseng exert beneficial effects on neurodegenerative diseases [10]. Ginseng suppresses oxidative stress, regulates apoptotic genes, and inhibits the toxicity of excitatory amino acids [11], [12]. Ginseng is classified into two groups: American ginseng and Asian ginseng. More than 38 ginsenosides, which are known as the major biologically active components, have been identified in Asian ginseng, and one of the major bioactive components is the ginsenoside Rg1 (Rg1) [13]. A number of studies have demonstrated the neuroprotective effects of Rg1 in neurodegenerative disorders such as AD [14]. It has been reported that β-secretase activity and Aβ-induced cytotoxicity is inhibited by Rg1 [15]. Although numerous studies have been conducted to assess the neuroprotective effects of Rg1, the protective mechanisms of Rg1 in AD remain elusive.
In the present study, we investigated the protective effects of Rg1 on SH-SY5Y cell damage induced by Aβ25–35. We performed an MS-based proteomic experiment using stable isotope labeling with amino acids in cell culture (SILAC) to explore the differential proteome expression associated with the protective effects of Rg1 in Aβ-treated neuronal cells. Our data may elucidate the neuroprotective molecular mechanism of Rg1 in AD.
2. Materials and methods
2.1. Aβ25–35 aggregation
Aβ25–35 (AnaSpec, Fremont, CA, USA) was dissolved in distilled water at a concentration of 2.5 mM and the solutions was incubated at 37°C for one week before use [16], [17].
2.2. SILAC
SILAC was performed as previously reported [18]. Briefly, SH-SY5Y cells were cultured for at least five cell divisions in either light SILAC medium containing 12C6-Arg and 12C6, 14N2-Lys or heavy SILAC medium containing 13C6-Arg and 13C6, 15N2-Lys supplemented with 10% dialyzed fetal bovine serum, 50 IU/mL penicillin, and 50 mg/mL streptomycin. The labeled cells were treated with (light medium) or without (heavy medium) 100μM Rg1 for 24 h in 25μM of Aβ25–35-treated cells for 24 h. The cells were lysed in buffer containing 1% Triton X-100, 150mM NaCl, 1mM EDTA, 50mM Tris-HCl (pH 8.0), 1mM sodium orthovanadate, 5mM NaF, 5mM sodium pyrophosphate, 1mM phenylmethylsulfonyl fluoride, aprotinin (1.5 μg/mL), antipain (10 mg/mL), leupeptin (10 μg/mL), and benzamidine (100 μg/mL). The lysates were centrifuged at 160,000g and mixed at a 1:1 protein ratio. The mixed lysates were separated by 10% SDS-PAGE and visualized with Coomassie Brilliant Blue G-250 (Bio-Rad, Hercules, CA, USA).
2.3. In-gel digestion
Each gel was cut into 10 bands of equal size, de-stained with 50% acetonitrile (ACN) in 25mM ammonium bicarbonate and dried in a speed vacuum concentrator. The dried gel pieces were re-swollen using 25mM ammonium bicarbonate (pH 8.0) containing 50 ng trypsin and incubated at 37°C for 24 h. The peptide mixtures were extracted with 50% ACN in 5% formic acid (FA) and dried in a speed vacuum concentrator.
2.4. MS analysis
MS was performed as previously described [18]. Briefly, the tryptic-dried peptides were analyzed using the Agilent HPLC-Chip/TOF MS system with the Agilent 1260 nano-LC system, HPLC Chip-cube MS interface and a 6530 QTOF single quadrupole-TOF mass spectrometer (Agilent Technologies, Santa Clara, CA, USA). The dried peptide samples were re-resolved in 2% ACN/0.1% FA and concentrated on a large-capacity HPLC Chip (Agilent Technologies). The HPLC chip incorporated an enrichment column (9 mm, 75 mm-inner diameter, 160 nL) and a reverse-phase column (15 cm, 75 mm-inner diameter, packed with Zorbax 300SB-C18 5 mm resins). The peptide separation was performed using a 70-min gradient of 3–45% buffer B (buffer A contained 0.1% FA/dry weight, and buffer B contained 90% ACN/0.1% FA/dry weight) at a flow rate of 300 nL/min. The MS and MS/MS data were acquired in positive ion mode and stored in centroid mode. The chip spray voltage was set at 1950 V and maintained under chip conditions. The drying gas temperature was set at 325°C with a flow rate of 3.5 L/min. A medium isolation (4 m/z) window was used for precursor isolation. A collision energy with a slope of 3.7 V/100 Da and an offset of 2.5 V were used for fragmentation. Additionally, while the MS data were acquired over a mass range of 300–3000 m/z, the MS/MS data were acquired over a mass range of 50–2500 m/z. Reference mass correction was performed using a reference mass of 922.0098. Precursors were set in an exclusion list for 0.5 min after two MS/MS spectra. The MS and MS/MS spectra were extracted using the Mass Hunter Qualitative Analysis B.05.00 software (Agilent Technologies) with default parameters. Database searches using the X! Tandem search engine were performed with a peptide mass tolerance of 20 ppm, an MS/MS tolerance of 0.5 Da, and a strict tryptic specificity (cleavage after lysine and arginine) allowing two missed cleavage sites; carbamidomethylation of Cys was set as a fixed modification, whereas oxidation (M) was considered a variable modification.
2.5. Protein validation and quantification
The MS database search results were analyzed using the Trans-Proteomic Pipeline (Systems Biology, Seattle, WA, USA), all assigned peptides were validated by PeptideProphet and the results filtered with an error rate of 0.05 (FDR of 5%) [19]. Peptide quantification was performed using the XPRESS modules included in the Trans-Proteomic Pipeline. Protein identification and quantification were validated using ProteinProphet and the result cutoff was set to a protein probability of 80%/error rate of 0.05 (FDR 5%) and a ratio of heavy from fixed to 1. SILAC ratios for proteins were quantified using XPRESS software [19]. The elution profiles of the light and heavy peptides were isolated and quantified based on the area of each peptide peak, and the abundance ratio was calculated based on these areas by XPRESS. Quantitative protein ratios were determined based on the average level of quantified peptides.
2.6. Bioinformatics analysis
Functional protein–protein network analysis was performed using the STRING web-based database Cytoscape plug-in [20]. The subcellular locations of the identified proteins were analyzed using the Uniprot database.
3. Results and discussion
To investigate the mechanisms associated with the neuroprotective activity of Rg1 in AD, we performed a comparative proteomic analysis of SH-SY5Y cells, which were treated with Rg1 after Aβ exposure using nano-LC-MS/MS combined with SILAC. We cultured SH-SY5Y cells in either light SILAC medium containing 12C6-Arg and 12C6, 14N2-Lys or heavy medium containing 13C6-Arg and 13C6, 15N2-Lys to label the cells with the stable isotopes [18]. Previous studies have shown that fresh and aged Aβ25–35 decreases the cell survival rate based on the results of the MTT reduction assay [21]. Therefore, we used aged Aβ25–35 to induce neuronal toxicity in SH-SY5Y cells. Cells cultured in light medium were exposed to aged Aβ25–35 and then treated with 100μM Rg1 [22], whereas cells cultured in heavy medium were treated with DMSO after exposure to aged Aβ25–35 (Fig. 1A).
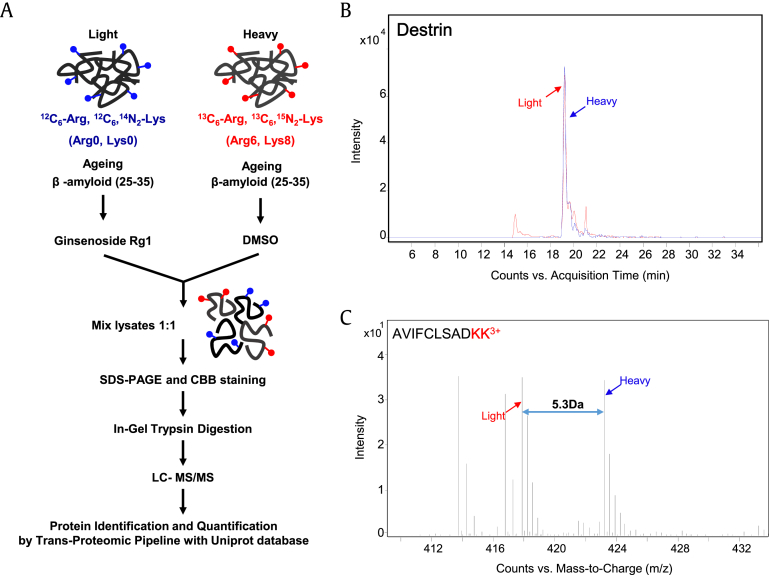
Fig. 1.
Stable isotope labeling with amino acids (SILAC) analysis of treatment with Rg1 in Aβ-treated cells. (A) Schematic diagram of the SILAC procedures used to quantify protein changes following treatment with Rg1 in β-amyloid peptide-treated SH-SY5Y cells. The lysate of cells cultured in SILAC medium was obtained, and the same amounts of proteins were combined and separated via SDS-PAGE followed by in-gel tryptic digestion. The tryptic peptides were analyzed by nano-LC MS/MS. (B) Paired extracted ion chromatograms and (C) monoisotopic mass peaks of destrin peptide (AVIFCLSADKK) from cells cultured in SILAC medium showed an approximate ratio of 1:1. The m/z difference between SILAC-labeled destrin peptides shows the triple charge of the destrin peptides.
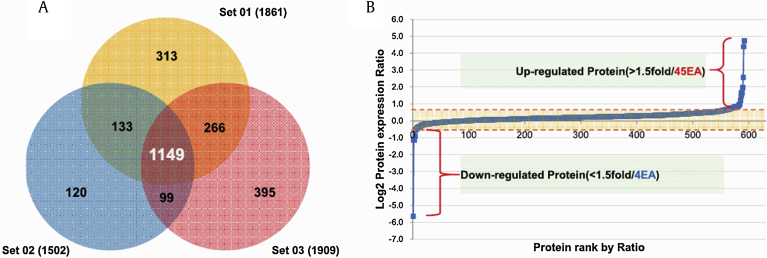
Three independent SILAC combined with LC-MS/MS experiments were performed. Fig. 1B shows a representative ion chromatogram of the AVIFCLSADKK peptide of destrin, and the monoisotopic peaks in the MS spectra of AVIFCLSADKK showed an expected 1:1 ratio (Fig. 1C). A total of 2,476 proteins were identified in two different physiological conditions. Of these, 1,149 proteins were common to the three data sets, and 592 proteins were quantified in at least two of the three sets (Fig. 2A and Table S1). Among the quantified proteins, 49 were significantly changed more than 1.5-fold; four proteins were downregulated and 45 proteins were upregulated in Aβ-treated SH-SY5Y cells exposed to Rg1 (Fig. 2B and Table 1).
Fig. 2.
Analysis of differentially expressed proteins following exposure of Aβ-treated cells to Rg1. (A) The Venn diagram shows the overall distribution of the proteins identified in three independent biological experiments. A total of 2,476 proteins were identified from three replicates, with 1,149 overlapping proteins. (B) Quantitative analysis of the proteins that were differentially expressed following the treatment of Aβ-treated cells with Rg1. Forty-five proteins were upregulated, and four proteins were significantly downregulated >1.5-fold.
Table 1.
List of proteins with a >1.5-fold change1)
| Accession No. | Gene name | Protein description | L/H log2 ratio |
|---|---|---|---|
| G3V2J8 | HSP90AA1 | Heat shock protein HSP 90-α (fragment) | −5.64 |
| O43432-3 | EIF4G3 | Isoform 3 of eukaryotic translation initiation factor 4 γ-3 | −1.12 |
| Q9Y3I0 | RTCB | tRNA-splicing ligase RtcB homolog | −1.12 |
| Q15369-2 | TCEB1 | Isoform 2 of Transcription elongation factor B polypeptide 1 | −0.92 |
| P18669 | PGAM1 | Phosphoglycerate mutase 1 | 0.58 |
| P52907 | CAPZA1 | F-actin-capping protein subunit α-1 | 0.58 |
| P37802-2 | TAGLN2 | Isoform 2 of transgelin-2 | 0.59 |
| P61970 | NUTF2 | Nuclear transport factor 2 | 0.59 |
| Q92616 | GCN1L1 | Translational activator GCN1 | 0.59 |
| O15144 | ARPC2 | Actin-related protein 2/3 complex subunit 2 | 0.61 |
| P08865 | RPSA | 40S ribosomal protein SA | 0.62 |
| O14880 | MGST3 | Microsomal glutathione S-transferase 3 | 0.63 |
| P04908 | HIST1H2AB | Histone H2A type 1-B/E | 0.64 |
| P84103-2 | SRSF3 | Isoform 2 of serine/arginine-rich splicing factor 3 | 0.64 |
| Q5JTZ9 | AARS2 | Alanine-tRNA ligase, mitochondrial | 0.64 |
| P08621-2 | SNRNP70 | Isoform 2 of U1 small nuclear ribonucleoprotein 70 kDa | 0.65 |
| P61086-2 | UBE2K | Isoform 2 of ubiquitin-conjugating enzyme E2 K | 0.65 |
| P62314 | SNRPD1 | Small nuclear ribonucleoprotein Sm D1 | 0.67 |
| Q07955-2 | SRSF1 | Isoform ASF-2 of serine/arginine-rich splicing factor 1 | 0.67 |
| Q99598 | TSNAX | Translin-associated protein X | 0.67 |
| Q15056-2 | EIF4H | Isoform short of eukaryotic translation initiation factor 4H | 0.69 |
| Q9UNF1 | MAGED2 | Melanoma-associated antigen D2 | 0.69 |
| Q7Z6Z7-2 | HUWE1 | Isoform 2 of E3 ubiquitin-protein ligase HUWE1 | 0.72 |
| P10412 | HIST1H1E | Histone H1.4 | 0.73 |
| P18206-2 | VCL | Isoform 1 of vinculin | 0.75 |
| Q96CN7 | ISOC1 | Isochorismatase domain-containing protein 1 | 0.75 |
| P51665 | PSMD7 | 26S proteasome non-ATPase regulatory subunit 7 | 0.76 |
| P60953 | CDC42 | Cell division control protein 42 homolog | 0.77 |
| P68036-2 | UBE2L3 | Isoform 2 of ubiquitin-conjugating enzyme E2 L3 | 0.77 |
| P38117-2 | ETFB | Isoform 2 of electron transfer flavoprotein subunit β | 0.79 |
| Q99714 | HSD17B10 | 3-hydroxyacyl-CoA dehydrogenase type-2 | 0.80 |
| P30153 | PPP2R1A | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A α isoform | 0.82 |
| P20674 | COX5A | Cytochrome c oxidase subunit 5A, mitochondrial | 0.84 |
| P28072 | PSMB6 | Proteasome subunit β type-6 | 0.85 |
| P47756-2 | CAPZB | Isoform 2 of F-actin-capping protein subunit β | 0.85 |
| O60888-2 | CUTA | Isoform A of protein CutA | 0.86 |
| Q14974 | KPNB1 | Importin subunit β-1 | 0.86 |
| P47914 | RPL29 | 60S ribosomal protein L29 | 0.86 |
| Q01518 | CAP1 | Adenylyl cyclase-associated protein 1 | 0.93 |
| Q99873-2 | PRMT1 | Isoform 2 of protein arginine N-methyltransferase 1 | 0.94 |
| Q9P1F3 | ABRACL | Costars family protein ABRACL | 0.98 |
| O14874-2 | BCKDK | Isoform 2 of [3-methyl-2-oxobutanoate dehydrogenase (lipoamide)] kinase, mitochondrial | 1.22 |
| P16949-2 | STMN1 | Isoform 2 of stathmin | 1.48 |
| P26583 | HMGB2 | High mobility group protein B2 | 1.66 |
| P31939-2 | ATIC | Isoform 2 of bifunctional purine biosynthesis protein PURH | 1.67 |
| P35268 | RPL22 | 60S ribosomal protein L22 | 1.98 |
| P13929-2 | ENO3 | Isoform 2 of β-enolase | 2.58 |
| A6NE09 | RPSAP58 | 40S ribosomal protein SA | 4.39 |
| O96008 | TOMM40 | Mitochondrial import receptor subunit TOM40 homolog | 4.75 |
Accession Nos. are from the Uniprot database. The fold change was calculated using Rg1 treated/control (unlabeled/labeled) ratios quantitated from integrated proteomics software. Ratios were obtained from n = 3
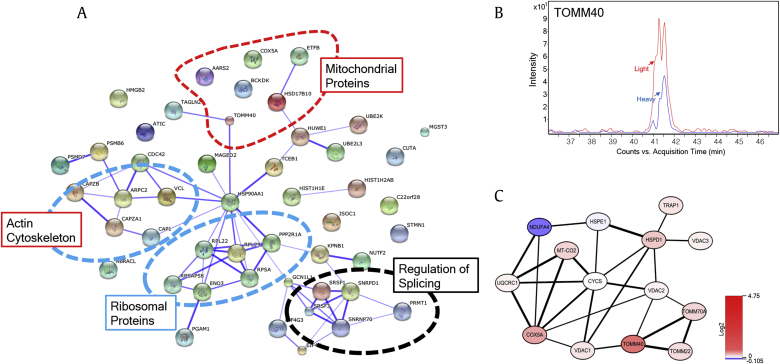
Next, we analyzed the protein–protein interaction networks among the proteins that were changed 1.5-fold using the STRING web-based protein database [20]. Protein–protein interaction clustering was distributed into four cellular locations: mitochondrial proteins (AARS2, ETFB, HSD17B10, COX5A, BCKDK, and TOMM40), actin cytoskeleton proteins (CAP1, CAPZA1, ARPC2, CAPZ2B, and VCL), ribosomal proteins (RPL22, RPL29, PPP2R1A, RPSAP58, ENO3, and RPSA), and regulation of splicing proteins (SRSF1, SNRPD1, SRSF3, SNRNP70, and PRMT1; Fig. 3A). Interestingly, we have previously reported that Rb1 is linked to the actin cytoskeleton in AD [18].
Fig. 3.
Protein–protein interaction analysis of proteins that were altered by treatment with Rg1 in Aβ-treated cells. (A) Protein–protein interactions were analyzed using the STRING web-based database. The proteins were grouped into four cellular locations: mitochondrial proteins, ribosomal proteins, actin cytoskeleton, and regulation of splicing proteins. (B) Extracted ion chromatogram of co-eluted heavy and light TOMM40 peptides forms. TOMM40 increased in Aβ-treated cells after treatment with Rg1. The average peptide heavy/light ratio was calculated from the ration of the peak area. (C) The protein interaction network analysis of the identified mitochondrial proteins was performed using the STRING database and visualized by Cytoscape (red, increase; blue, decrease).
We noted the identification of mitochondrial proteins because a number of studies have shown that mitochondrial dysfunction is associated with the pathogenesis of AD [3]. Interestingly, the translocase of the outer membrane (TOM) complex has been reported to be the main entrance for most cytoplasmically synthesized mitochondrial proteins, and a decrease in TOMM40 levels causes mitochondrial dysfunction associated with neurodegenerative diseases such as AD [23]. Furthermore, the expression level of TOMM40 was found to be significantly downregulated in blood samples of patients with AD [24], [25]. By contrast, our data showed the opposite results following the treatment of Aβ-treated SH-SY5Y cells with Rg1. Fig. 3B shows the relative quantification of TOMM40 expression using an ion chromatogram from MS, which showed that TOMM40 was dramatically increased following treatment with Rg1.
Our previous study showed that mitochondrial proteins were dramatically altered in MPP+-induced neuronal cells based on a quantitative proteomic analysis. Furthermore, the mitochondrial proteins HSD17B10 and AARS2 containing TOMM40 were dramatically downregulated in response to MPP+-induced neuronal damage, which is known to be associated with neurodegeneration [19], [26]. However, our results showed that HSD17B10 and AARS2 were noticeably increased by the treatment with Rg1 in Aβ-treated SH-SY5Y cells (Table 1). These data indicate that Rg1 might have a protective role in the regulation of mitochondrial functions in AD.
To further investigate the functional role of Rg1 in relation to mitochondrial proteins in AD, all of the mitochondrial proteins identified in this study were further explored in the available literature. We found 14 mitochondrial proteins that have been reported to be associated with AD (Table S2), and we analyzed the protein–protein interaction network among the mitochondrial proteins using the STRING database plug-in Cytoscape. Fig. 3C shows that 14 mitochondrial proteins exhibited a strong interaction network. The thickness of the black connection lines indicates the protein–protein interaction score. Interestingly, VDAC1 has been shown to be significantly increased in AD and transgenic mice [27], [28], and abnormally high levels of Aβ interact with VDAC and interrupt metabolite transport via pore blockade [29]. VDAC1 protein expression was not altered by Rg1 treatment after Aβ exposure, in contrast to previous results showing increased levels of VDAC1 in AD. COX5A is the terminal oxidase of the mitochondrial electron transport chain and binds to Aβ, resulting in a malfunction of COX5A in cellular energy metabolism in AD and amyloid precursor protein transgenic mice [30]. Interestingly, the interaction between COX5A and TOMM40 is mediated by VDAC1, and the expression of these proteins exhibits a pattern opposite to that observed in Rg1-treated AD (Fig. 3C). A previous study has shown that NDUFA4 is highly expressed in AD [31]. However, Rg1 treatment decreased NDUFA4. Taken together, our data suggest that Rg1 might play a critical role in regulating the functions of mitochondrial proteins in AD.
In conclusion, numerous studies have been performed to investigate the potential mechanisms of ginsenosides in neuronal diseases such as AD. To explore the neuronal protective mechanism of Rg1 in AD, we performed a global proteomic analysis using SILAC together with LC-MS/MS. In total, 1,149 proteins were identified in the three replicated experiments. A total of 45 proteins showed significant alterations in response to Rg1 treatment in Aβ-pretreated cells. Our data showed that these proteins were associated with mitochondrial proteins, actin cytoskeleton proteins, ribosomal proteins, and proteins that regulate splicing. In particular, the results showed that mitochondrial proteins containing HSD17B10, AARS2, TOMM40, VDAC1, COX5A, and NDUFA4 might be related to the protective mechanisms of Rg1 in AD. Additional studies are needed to investigate the detailed molecular mechanisms that link Rg1 to these proteins in AD.
Conflicts of interest
All authors have no conflicts of interest to declare.
Acknowledgments
This work was performed with the support of the Cooperative Research Program for Agriculture Science & Technology Development (PJ00983503), Rural Development Administration, Republic of Korea, and the National Research Foundation of Korea Grants NRF-2015R1A2A2A01006965.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.jgr.2016.12.003.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Correa M.S., Vedovelli K., Giacobbo B.L., de Souza C.E., Ferrari P., de Lima Argimon I.I., Walz J.C., Kapczinski F., Bromberg E. Psychophysiological correlates of cognitive deficits in family caregivers of patients with Alzheimer disease. Neuroscience. 2015;286:371–382. doi: 10.1016/j.neuroscience.2014.11.052. [DOI] [PubMed] [Google Scholar]
- 2.Mucke L. Neuroscience: Alzheimer's disease. Nature. 2009;461:895–897. doi: 10.1038/461895a. [DOI] [PubMed] [Google Scholar]
- 3.Piaceri I., Rinnoci V., Bagnoli S., Failli Y., Sorbi S. Mitochondria and Alzheimer's disease. J Neurol Sci. 2012;322:31–34. doi: 10.1016/j.jns.2012.05.033. [DOI] [PubMed] [Google Scholar]
- 4.Selkoe D.J., Schenk D. Alzheimer's disease: molecular understanding predicts amyloid-based therapeutics. Annu Rev Pharmacol Toxicol. 2003;43:545–584. doi: 10.1146/annurev.pharmtox.43.100901.140248. [DOI] [PubMed] [Google Scholar]
- 5.Masters C.L., Simms G., Weinman N.A., Multhaup G., McDonald B.L., Beyreuther K. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc Natl Acad Sci U S A. 1985;82:4245–4249. doi: 10.1073/pnas.82.12.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vassar R., Bennett B.D., Babu-Khan S., Kahn S., Mendiaz E.A., Denis P., Teplow D.B., Ross S., Amarante P., Loeloff R. Beta-secretase cleavage of Alzheimer's amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- 7.Pinho C.M., Teixeira P.F., Glaser E. Mitochondrial import and degradation of amyloid-beta peptide. Biochim Biophys Acta. 2014;1837:1069–1074. doi: 10.1016/j.bbabio.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 8.Iversen L.L., Mortishire-Smith R.J., Pollack S.J., Shearman M.S. The toxicity in vitro of beta-amyloid protein. Biochem J. 1995;311:1–16. doi: 10.1042/bj3110001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reddy P.H., Beal M.F. Amyloid beta, mitochondrial dysfunction and synaptic damage: implications for cognitive decline in aging and Alzheimer's disease. Trends Mol Med. 2008;14:45–53. doi: 10.1016/j.molmed.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho I.H. Effects of Panax ginseng in neurodegenerative diseases. J Ginseng Res. 2012;36:342–353. doi: 10.5142/jgr.2012.36.4.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen X.C., Zhu Y.G., Zhu L.A., Huang C., Chen Y., Chen L.M., Fang F., Zhou Y.C., Zhao C.H. Ginsenoside Rg1 attenuates dopamine-induced apoptosis in PC12 cells by suppressing oxidative stress. Eur J Pharmacol. 2003;473:1–7. doi: 10.1016/s0014-2999(03)01945-9. [DOI] [PubMed] [Google Scholar]
- 12.Shen L., Zhang J. NMDA receptor and iNOS are involved in the effects of ginsenoside Rg1 on hippocampal neurogenesis in ischemic gerbils. Neurol Res. 2007;29:270–273. doi: 10.1179/016164107X159144. [DOI] [PubMed] [Google Scholar]
- 13.Im D.S., Nah S.Y. Yin and Yang of ginseng pharmacology: ginsenosides vs gintonin. Acta Pharmacol Sin. 2013;34:1367–1373. doi: 10.1038/aps.2013.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li N., Zhou L., Li W., Liu Y., Wang J., He P. Protective effects of ginsenosides Rg1 and Rb1 on an Alzheimer's disease mouse model: a metabolomics study. J Chromatogr B Analyt Technol Biomed Life Sci. 2015;985:54–61. doi: 10.1016/j.jchromb.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y.H., Du G.H. Ginsenoside Rg1 inhibits beta-secretase activity in vitro and protects against Abeta-induced cytotoxicity in PC12 cells. J Asian Nat Prod Res. 2009;11:604–612. doi: 10.1080/10286020902843152. [DOI] [PubMed] [Google Scholar]
- 16.Giovannelli L., Scali C., Faussone-Pellegrini M.S., Pepeu G., Casamenti F. Long-term changes in the aggregation state and toxic effects of beta-amyloid injected into the rat brain. Neuroscience. 1998;87:349–357. doi: 10.1016/s0306-4522(98)00169-9. [DOI] [PubMed] [Google Scholar]
- 17.Giovannelli L., Casamenti F., Scali C., Bartolini L., Pepeu G. Differential effects of amyloid peptides beta-(1-40) and beta-(25-35) injections into the rat nucleus basalis. Neuroscience. 1995;66:781–792. doi: 10.1016/0306-4522(94)00610-h. [DOI] [PubMed] [Google Scholar]
- 18.Hwang J.Y., Shim J.S., Song M.Y., Yim S.V., Lee S.E., Park K.S. Proteomic analysis reveals that the protective effects of ginsenoside Rb1 are associated with the actin cytoskeleton in beta-amyloid-treated neuronal cells. J Ginseng Res. 2016;40:278–284. doi: 10.1016/j.jgr.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi J.W., Song M.Y., Park K.S. Quantitative proteomic analysis reveals mitochondrial protein changes in MPP(+)-induced neuronal cells. Mol Biosyst. 2014;10:1940–1947. doi: 10.1039/c4mb00026a. [DOI] [PubMed] [Google Scholar]
- 20.Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shearman M.S., Ragan C.I., Iversen L.L. Inhibition of PC12 cell redox activity is a specific, early indicator of the mechanism of beta-amyloid-mediated cell death. Proc Natl Acad Sci U S A. 1994;91:1470–1474. doi: 10.1073/pnas.91.4.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saw C.L., Yang A.Y., Cheng D.C., Boyanapalli S.S., Su Z.Y., Khor T.O., Gao S., Wang J., Jiang Z.H., Kong A.N. Pharmacodynamics of ginsenosides: antioxidant activities, activation of Nrf2, and potential synergistic effects of combinations. Chem Res Toxicol. 2012;25:1574–1580. doi: 10.1021/tx2005025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gottschalk W.K., Lutz M.W., He Y.T., Saunders A.M., Burns D.K., Roses A.D., Chiba-Falek O. The broad impact of TOM40 on neurodegenerative diseases in aging. J Parkinsons Dis Alzheimers Dis. 2014 doi: 10.13188/2376-922X.1000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee T.S., Goh L., Chong M.S., Chua S.M., Chen G.B., Feng L., Lim W.S., Chan M., Ng T.P., Krishnan K.R. Downregulation of TOMM40 expression in the blood of Alzheimer disease subjects compared with matched controls. J Psychiatr Res. 2012;46:828–830. doi: 10.1016/j.jpsychires.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 25.Goh L.K., Lim W.S., Teo S., Vijayaraghavan A., Chan M., Tay L., Ng T.P., Tan C.H., Lee T.S., Chong M.S. TOMM40 alterations in Alzheimer's disease over a 2-year follow-up period. J Alzheimers Dis. 2015;44:57–61. doi: 10.3233/JAD-141590. [DOI] [PubMed] [Google Scholar]
- 26.Yang S.Y., He X.Y., Schulz H. 3-Hydroxyacyl-CoA dehydrogenase and short chain 3-hydroxyacyl-CoA dehydrogenase in human health and disease. FEBS J. 2005;272:4874–4883. doi: 10.1111/j.1742-4658.2005.04911.x. [DOI] [PubMed] [Google Scholar]
- 27.Reddy P.H. Is the mitochondrial outermembrane protein VDAC1 therapeutic target for Alzheimer's disease? Biochim Biophys Acta. 2013;1832:67–75. doi: 10.1016/j.bbadis.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ren R., Zhang Y., Li B., Wu Y., Li B. Effect of beta-amyloid (25-35) on mitochondrial function and expression of mitochondrial permeability transition pore proteins in rat hippocampal neurons. J Cell Biochem. 2011;112:1450–1457. doi: 10.1002/jcb.23062. [DOI] [PubMed] [Google Scholar]
- 29.Manczak M., Reddy P.H. Abnormal interaction of VDAC1 with amyloid beta and phosphorylated tau causes mitochondrial dysfunction in Alzheimer's disease. Hum Mol Genet. 2012;21:5131–5146. doi: 10.1093/hmg/dds360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hernandez-Zimbron L.F., Luna-Muñoz J., Mena R., Vazquez-Ramirez R., Kubli-Garfias C., Cribbs D.H., Manoutcharian K., Gevorkian G. Amyloid-beta peptide binds to cytochrome C oxidase subunit 1. PLoS One. 2012;7:e42344. doi: 10.1371/journal.pone.0042344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sekar S., McDonald J., Cuyugan L., Aldrich J., Kurdoglu A., Adkins J., Serrano G., Beach T.G., Craig D.W., Valla J. Alzheimer's disease is associated with altered expression of genes involved in immune response and mitochondrial processes in astrocytes. Neurobiol Aging. 2015;36:583–591. doi: 10.1016/j.neurobiolaging.2014.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.