Many organisms rely on active, directed cell movement for their survival and/or propagation. Unicellular organisms and specialized cells of metazoans rely on cell movement for essential tasks such as gamete pairing, predator evasion, and the search for prey. Although many types of cell motility exist, they can be divided into 2 broad classes representing the 2 main physical environments encountered by cells: swimming through liquid and crawling across and between solid surfaces.
Apart from some notable exceptions, the majority of swimming eukaryotic cells rely on whip-like, microtubule-based organelles called flagella. It is now generally accepted that the flagellum was present in the last common ancestor of all living eukaryotes and has been retained by members of all major lineages.1 There are 2 distinct lines of evidence for this single origin of flagella. The first to be uncovered was the conservation of intricate substructures, revealed by electron micrographs of flagella from diverse organisms. This structural evidence was later supported by molecular phylogenies of flagellar proteins that supported the concept of a single evolutionary origin.1 Genes encoding many proteins found only inside flagella have been retained only in those organisms that build flagella. This finding indicates that organisms that do not build flagella have lost this capacity as well as the genes required for it.1,2
These 2 lines of evidence—the conservation of flagellar substructure and the genes unique to flagella—make a convincing case for a single evolutionary origin of flagellar-based swimming. In contrast, the origin of crawling motility remains mysterious.3 To trace its evolutionary history, we must first identify cellular structures used for crawling motility, and determine whether their substructures and components suggest a common origin or an evolutionary convergence.
Unlike eukaryotic swimming, which uses a single, well defined and stable organelle (the flagellum), cells can crawl using a variety of morphologically distinct structures, many of which change shape rapidly and cyclically assemble and disassemble during the process of migration.4 The best studied structures used for cell crawling are the focal adhesions used by fibroblasts, epithelial cells, and other highly adherent animal cells to grip molecules of the extracellular matrix to pull themselves along. Other cellular structures are used by a wider range of crawling cells (Fig. 1).4 A widely-used mode of crawling uses the extension of broad protrusions filled with networks of branched actin filaments. These “pseudopods” are used by single-cell amoebae as well as some white blood cells. Other organisms appear to use linear “filopodia,” some of which are possibly filled with bundled actin filaments, to walk across a surface. In addition to these actin-filled protrusions, many cell types and organisms can also move forward by popping out spherical, cytoplasm-filled protrusions called blebs.
Figure 1.
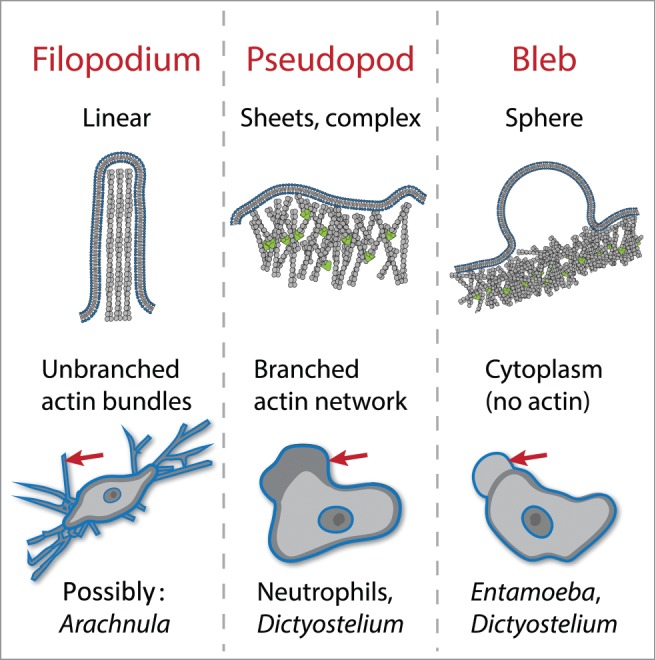
Actin-dependent structures used for eukaryotic cell crawling. Filopodia are linear protrusions formed by elongation of bundled actin filaments. Pseudopods are broad protrusions filled with branched actin networks that push out the membrane. Blebs are cytoplasm-filled spheres formed by delamination of the membrane from the underlying actin cortex. Examples of cells that use each for motility are listed. Many cells also use these protrusions for feeding and probing the environment.
Each of these modes of crawling depends on distinct molecular mechanisms, and we must trace the evolution of each independently. We currently focus on actin-driven, pseudopod-based crawling because of the important role it plays in the function of both medically important white blood cells and many heterotrophs occupying key niches of ecological food webs. We have begun to test the hypothesis that, like eukaryotic flagella, branched-actin-filled pseudopods share a single evolutionary origin.3 To this end, we parallel the strategies used to investigate the evolutionary history of flagella, asking whether pseudopods built by evolutionarily diverse cells share the same substructures, and then identifying and tracing the evolutionary history of molecular components of pseudopods.
Looking more closely at pseudopods using electron microscopy as well as immunofluorescence reveals that these dynamic protrusions are filled with growing branched actin networks5 (Fig. 1). Similar actin networks are also used for a variety of other cell processes, including both endocytosis and cytokinesis.5 To date, electron microscopy has not revealed a regular structure that would differentiate it from the branched-actin networks used for a variety of other cell processes. Therefore, unlike flagellar substructure, pseudopod substructure provides no clues as to whether they evolved once, or whether branched actin networks were co-opted for pseudopod assembly multiple times during eukaryotic history.
We are therefore left with tracing the evolutionary history of genes used specifically to build and regulate pseudopods to understand how and when this mode of cell crawling arose. Multi-purpose components, such as actin itself, are not helpful because they are required for a wide variety of cellular processes. To date, the only molecular signature that appears to differentiate the actin networks used for pseudopods from those used for other cell processes that use branched actin is the pseudopod's reliance on 2 upstream activators of actin assembly, WASP and SCAR/WAVE. As we recently showed,3 both proteins are co-conserved in species that build pseudopods, both are needed to assemble robust pseudopods by human neutrophils, and the presence of both genes can actually predict the ability to form pseudopods by little-studied species. These findings are the first evidence to support a single evolutionary origin of branched-actin pseudopod-based cell motility. However, this single piece of evidence is insufficient proof, and we are currently testing whether the function of other genes that appear to be co-conserved with WASP and SCAR/WAVE can shed more light on the evolutionary history of pseudopod-based cell crawling. We hope we and others can use similar approaches to understanding the origin of other modes of crawling, as well as other complex cellular behaviors.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Carvalho-Santos Z, Azimzadeh J, Pereira-Leal JB, Bettencourt-Dias M. Evolution: tracing the origins of centrioles, cilia, and flagella. J Cell Biol. 2011;194:165-75. doi: 10.1083/jcb.201011152. PMID:21788366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Fritz-Laylin LK, Prochnik SE, Ginger ML, Dacks JB, Carpenter ML, Field MC, Kuo A, Paredez A, Chapman J, Pham J, Shu S, Neupane R, Cipriano M, Mancuso J, Tu H, Salamov A, Lindquist E, Shapiro H, Lucas S, Grigoriev IV, Cande WZ, Fulton C, Rokhsar DS, Dawson SC.. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell. 2010;140:631-42. doi: 10.1016/j.cell.2010.01.032. PMID:20211133 [DOI] [PubMed] [Google Scholar]
- [3].Fritz-Laylin LK, Lord SJ, Mullins RD. WASP and SCAR are evolutionarily conserved in actin-filled pseudopod-based motility. J Cell Biol. 2017;216:1673-88. doi: 10.1083/jcb.201701074. PMID:28473602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lämmermann T, Sixt M. Mechanical modes of “amoeboid” cell migration. Curr Opin Cell Biol. 2009;21:636-44. doi: 10.1016/j.ceb.2009.05.003. PMID:19523798 [DOI] [PubMed] [Google Scholar]
- [5].Svitkina TM. Ultrastructure of protrusive actin filament arrays. Curr Opin Cell Biol. 2013;25:574-81. doi: 10.1016/j.ceb.2013.04.003. PMID:23639311 [DOI] [PMC free article] [PubMed] [Google Scholar]


