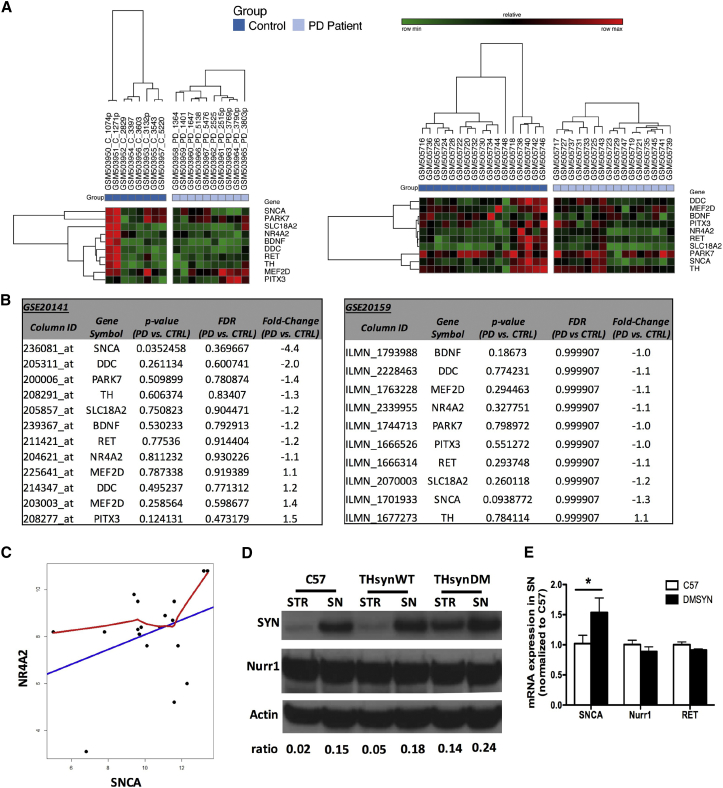
Figure 1.
Nurr1 and Associated Genes Expression in Sporadic PD Patients and α-syn Transgenic Mice
(A) Hierarchical clustering heatmaps for microarray datasets GEO: GSE20141 (left panel) and GSE20159 (right panel). (B) Expression of genes of interest in the microarray datasets (GEO: GSE20141, left table; GEO: GSE20159, right table). The p values as well as Benjamini-Hochberg false discovery rates (FDRs) and fold change were reported for every single gene. (C) The scatterplot was drawn from the robust multi-array average (RMA)-processed and logarithm 2-transformed microarray intensity data of 18 samples. The x axis represents SNCA as an independent variable, and the y axis represents NR4A2 as a dependent variable. The straight blue line is the linear regression result between the two genes SNCA and NR4A2, and the red line is gained by the local weighted scatterplot smoothing (LOWESS) regression analysis. The correlation coefficient is 0.4084 with p = 0.1442. (D) α-syn and Nurr1 protein expression in the striatum and nigra of C57, THsynWT, and THsynDM mice. Striatum (STR) and nigra (SN) tissues were microdissected from 6-month-old transgenic mice with expression of wild-type synuclein (THsynWT) or doubly mutated α-syn (A30P & A53T; THsynDM) or age-controlled C57. The expression of α-syn and Nurr1 was determined by western blot analysis. The signal density ratio of α-syn over action was quantified. (E) α-syn, Nurr1, and RET gene expression in the SN were determined by qRT-PCR.