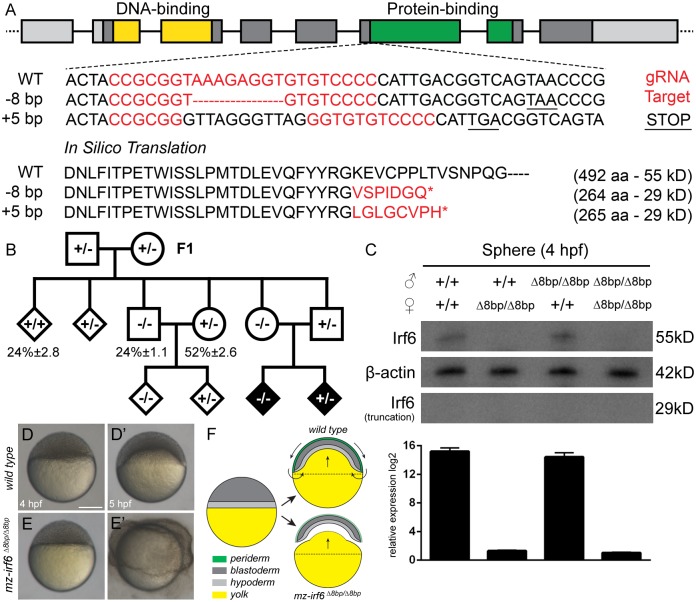
Fig 1. Generation and characterization of the zebrafish irf6 CRISPR mutant model.
(A) Zebrafish irf6 gene structure composed of eight exons, a helix-turn-helix DNA-binding domain (yellow), and a SMIR/IAD protein-binding domain (green). The CRISPR gRNA target site was located in exon 6 at the start of the protein-binding domain. Sanger sequencing of the target site revealed a -8bp deletion (Δ8bp) that created a frameshift and premature stop codon, truncating the protein to 29 kD as predicted by in silico translation. Another +5bp insertion was also identified. (B) Breeding pedigree revealed the irf6 mutant phenotype in F3 and the importance of maternal transcripts. (C) Top: western blot at the sphere stage (4 hpf) revealed a lack of Irf6 full-length (55 kD) or truncated (29 kD) protein in all maternal irf6 Δ8bp/Δ8bp embryos but not paternal irf6 Δ8bp/Δ8bp embryos or wild type embryos. Bottom: relative gene expression by RT-qPCR revealed a lack of irf6 mRNA transcripts in all maternal irf6 Δ8bp/Δ8bp embryos but comparable levels between wild type and paternal irf6 Δ8bp/Δ8bp embryos. Error bar = 2xSEM, n = 3. (D-D’) Wild type embryos at the sphere stage (4 hpf) (D) and at the 30% epiboly stage (5 hpf) (D’). (E-E’) Maternal irf6 Δ8bp/Δ8bp embryos at the sphere stage (4 hpf) (E) and displaying the periderm rupture phenotype at 5 hpf (E’). Scale bar = 250 μm. (F) Cross-sectional schematic through the embryonic midline illustrating the zebrafish embryo epiboly process. Arrows represent cell and yolk directional movements. Wild type embryos experience rapid cellular lamination and yolk doming between 4–5 hpf, while maternal-zygotic irf6 Δ8bp/Δ8bp embryos experience incomplete periderm differentiation and animal pole/yolk separation.