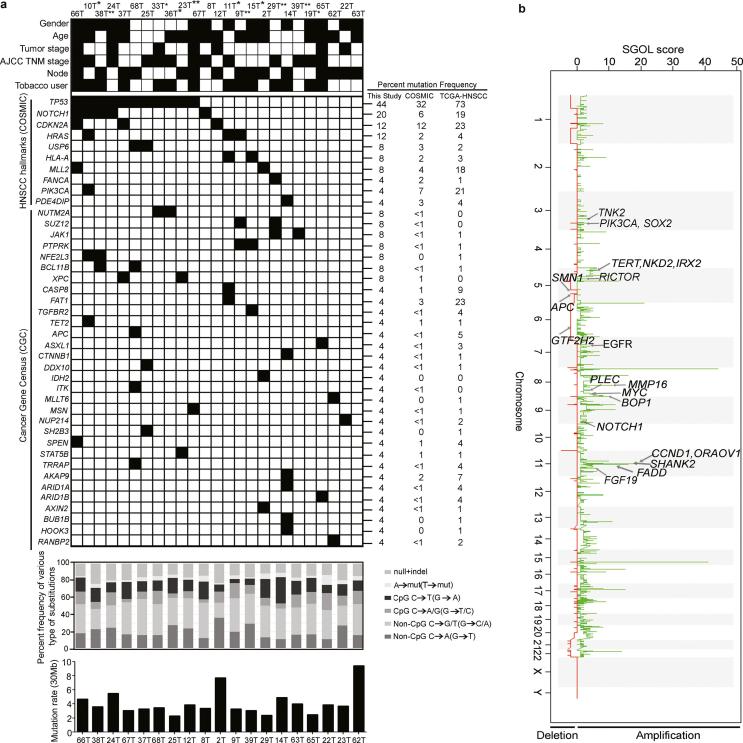
Fig. 1.
Identification of somatic mutations and DNA copy number changes in HPV-negative early stage TSCC tumors. (a) Mutational features of 25 early tongue squamous carcinoma samples: 19 of 24 whole exome (5 samples excluded due to low quality reads) and 6 of 11 whole transcriptome sequencing (excluding 5 common samples). Samples ID’s with asterisk (**) denotes samples with exome and transcriptome sequencing; (*) samples with transcriptome sequencing alone. Different clinicopathological factors such as; gender, age, tumor stage, AJCC TNM stage and lymph node metastasis status and etiological factors such as tobacco users are shown for each patient. The black solid boxes denotes gender: male, age: >45 years, tumor stage: pT1, AJCC-TNM stage: Stage I–II, nodal status: positive and tobacco habit. The white boxes denote gender: female, age: <45 years, tumor stage: pT2, AJCC-TNM stage: Stage III–IV, nodal status: negative and without tobacco habit. Grey filled boxes denotes no information available. The ten HNSCC hallmark genes and cancer gene census (COSMIC) found to be mutated in data, is arranged in decreasing order of percent frequency. Black filled box denotes presence of a somatic mutation in the patient. Mutation frequencies for the hallmark and cancer census genes observed in this study (n = 25), COSMIC-HNSCC (n ≥ 500) and TCGA-HNSCC (n = 279) samples. The substitution frequencies spectrum for each patient for whole exome sequencing data is shown. Percent frequency of various types of SNVs and indels are shown. Different types of substitutions shown by different shades. Somatic non-silent mutation rate/30 Mb derived from whole exome sequencing data for each tumor is shown. (b) Somatic DNA copy number changes identified using Exome sequencing data. Somatic DNA copy number gains and losses were generated using Segments-of-Gain-Or-Loss (SGOL) scores across 23 TSCC patients. SGOL score is plotted (horizontal axis) for DNA copy number gains (green) and losses (red) are plotted as a function of distance along with human genome (vertical axis). Representative amplified and deleted regions are annotated for HNSCC-associated genes and denoted by an arrow. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)