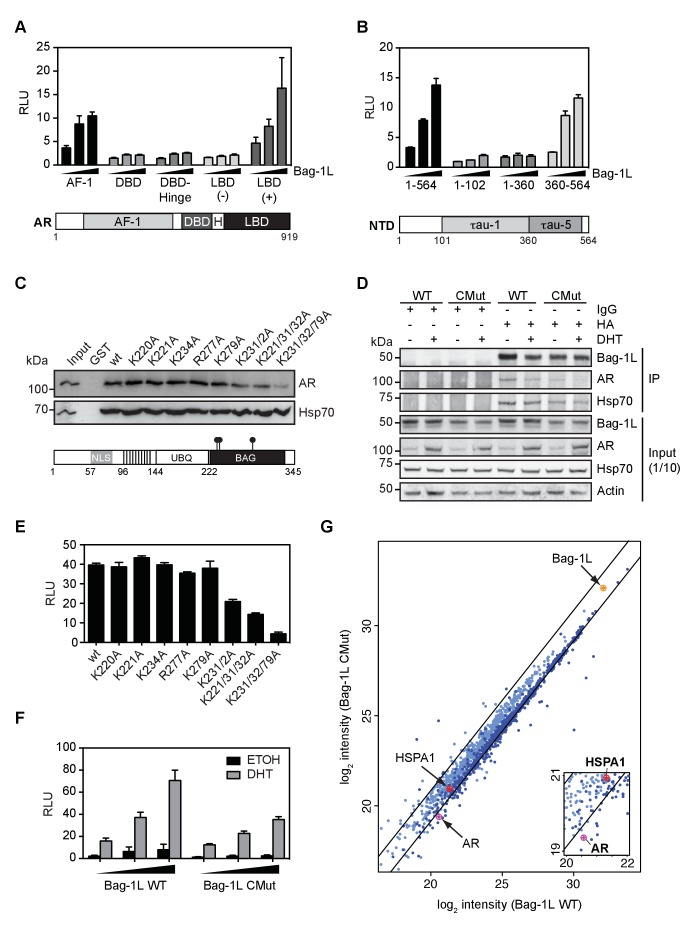
Figure 2. The Bag-1L:AR interaction is mediated by K231/232/279 in the BAG domain of Bag-1L.
(A, B) Mammalian one-hybrid assay in HeLa cells transfected with indicated AR domains linked to Gal4 DBD, subjected to increasing concentration of Bag-1L. The results are the mean of three independent experiments ± SEM, relative to the empty Bag-1L expression vector. Schematic representations of the AR domains are shown below. AF-1: Activation function-1; H: Hinge; DBD: DNA-binding domain; LBD: Ligand-binding domain. (C) GST pull-down with GST-Bag-1L fusion proteins harboring point mutations (as indicated) in their BAG domain and lysates from LNCaP cells. Shown below is a schematic structure of Bag-1L with the triple mutations in the BAG domain, which abolish the interaction with the AR (but have no effect on Hsp70 binding). NLS: Nuclear localization sequence; UBQ: Ubiquitin-like domain; BAG: BAG domain. (D) Co-immunoprecipitation of Bag-1L and AR in LNCaP cells stably overexpressing FLAG-, HA-tagged wild-type (WT) or BAG domain mutant Bag-1L (CMut). The IP was performed using an anti-HA-tag antibody against Bag-1L and an antibody against AR and Hsp70 to evaluate binding of these proteins to Bag-1L. Equal protein loading was confirmed by probing for expression of Bag-1L, AR, Hsp70 and β-actin.. (E) Mammalian one-hybrid assay in HeLa cells transfected with pG5ΔE4-38 luciferase, TK Renilla luciferase, pM-AR AF-1 and different Bag-1L constructs harboring a wild-type or mutant BAG domain (as indicated). The results are the mean of three independent experiments ± SEM, relative to the empty Bag-1L expression vector. (F) Mammalian two-hybrid assay in HeLa cells transfected with Gal4 DBD-AR LBD and VP16-AR-AF-1 and increasing amounts of wild-type (WT) or K231/232/279A mutant Bag-1L (CMut). The results are the mean of three independent experiments ± SEM, relative to the control Renilla luciferase. (G) Log-log plot of intensities for proteins detected in forward and reverse SILAC RIME analyses of Bag-1L WT and CMut cells, targeting BAG-1L (dark blue) or IgG (light blue). Black lines represent median IgG-RIME ratios ± 2 standard deviations. Bag-1L, Hsp70 (HSPA1) and AR are indicated in yellow and red, respectively.
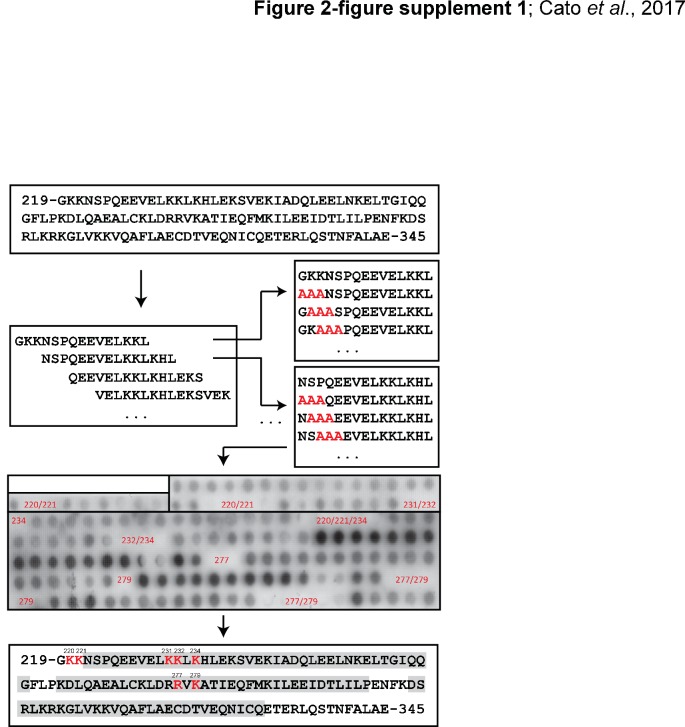
Figure 2—figure supplement 1. Schematic of the SPOT synthesis technology.
Figure 2—figure supplement 2. Overlap between AR cistromes in wild-type and CMut Bag-1L-expressing LNCaP cells.
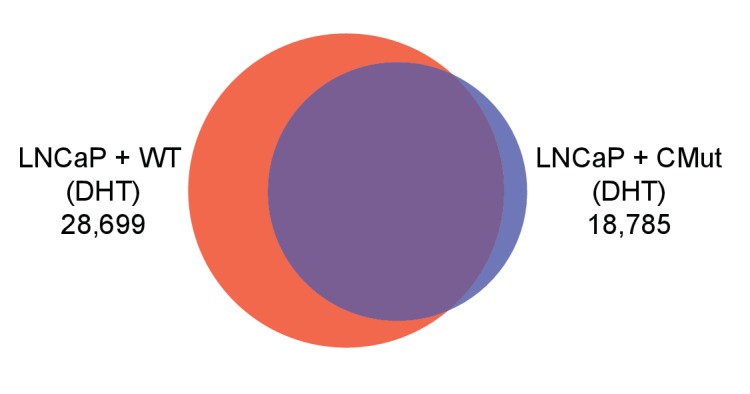
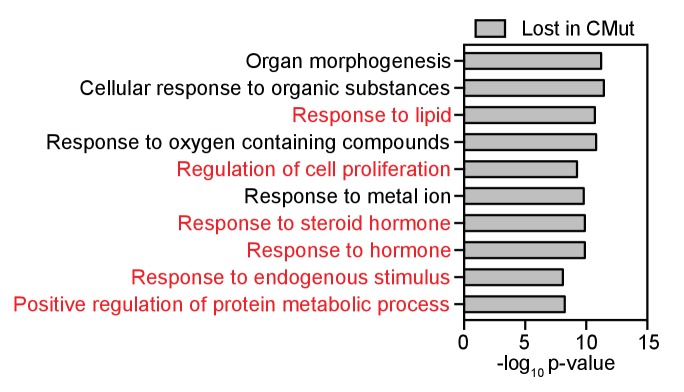
Figure 2—figure supplement 3. Top ten GO-terms (GSEA) associated with direct AR-target genes lost in the Bag-1L CMut- compared to the wild-type Bag-1L-expressing cells.
Figure 2—figure supplement 4. Conserved BAG domain mutations that inhibit the AR AF-1 transactivation.
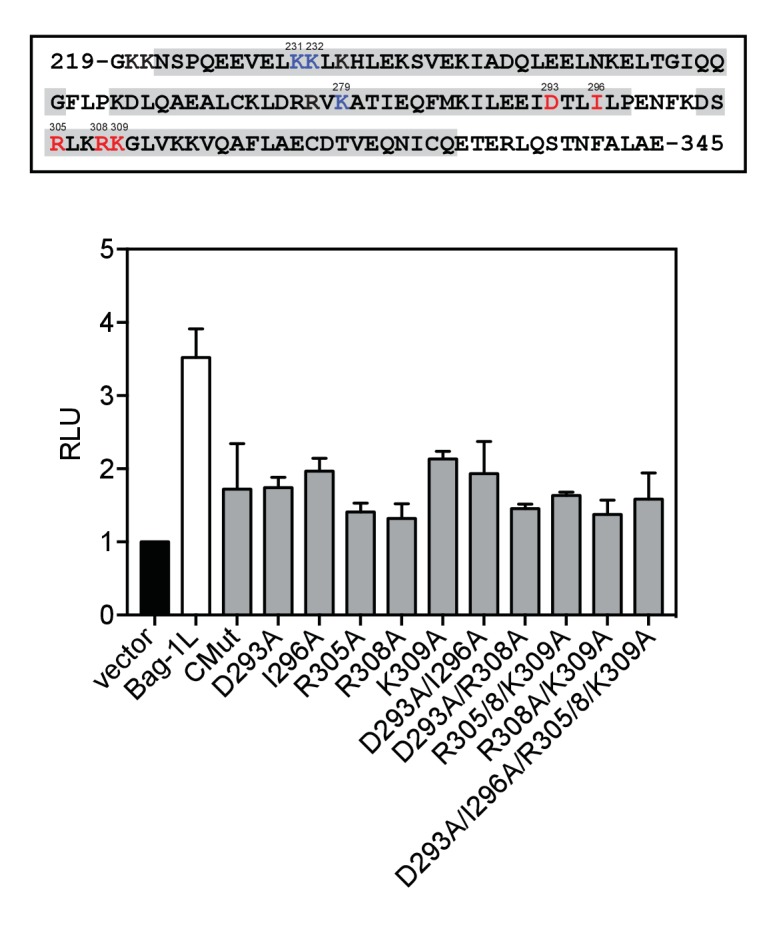
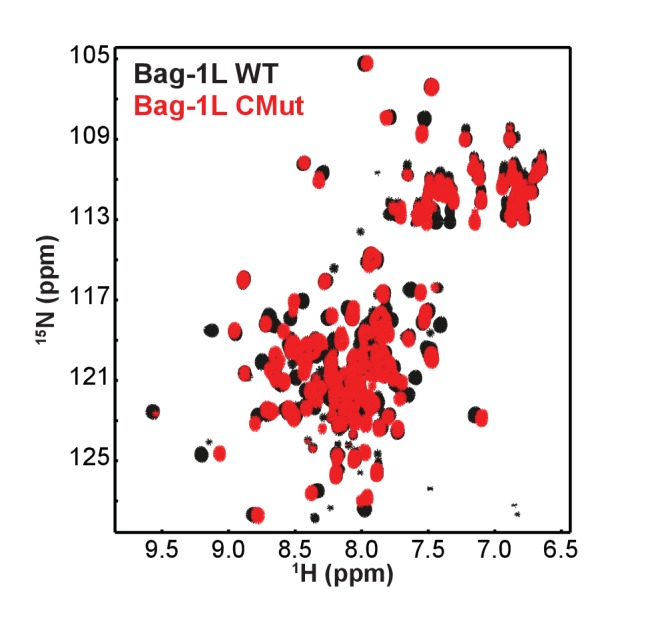
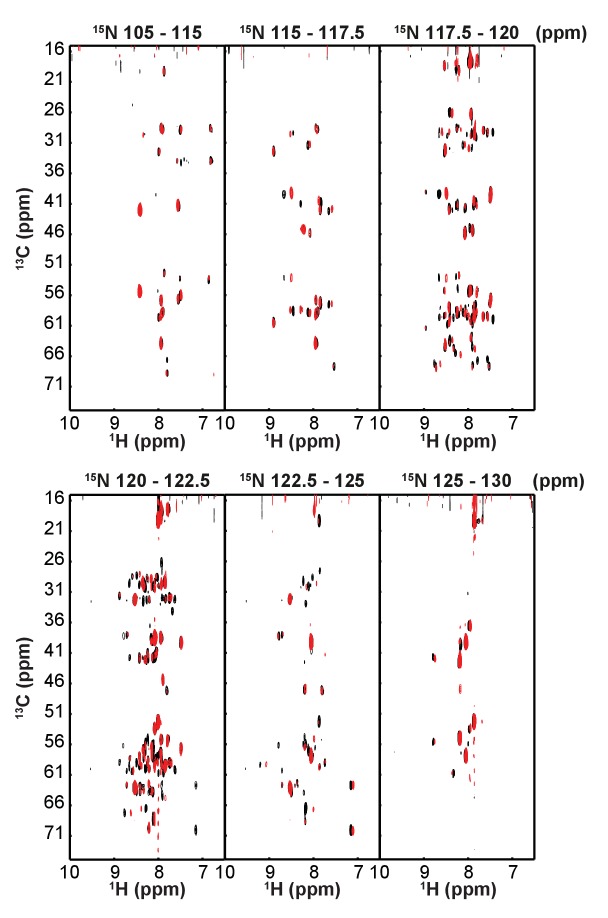
Figure 2—figure supplement 5. CBCACONH data of wild-type and CMut Bag-1L.
Figure 2—figure supplement 6. 15N-HSQC spectra of wild-type and CMut Bag-1L.