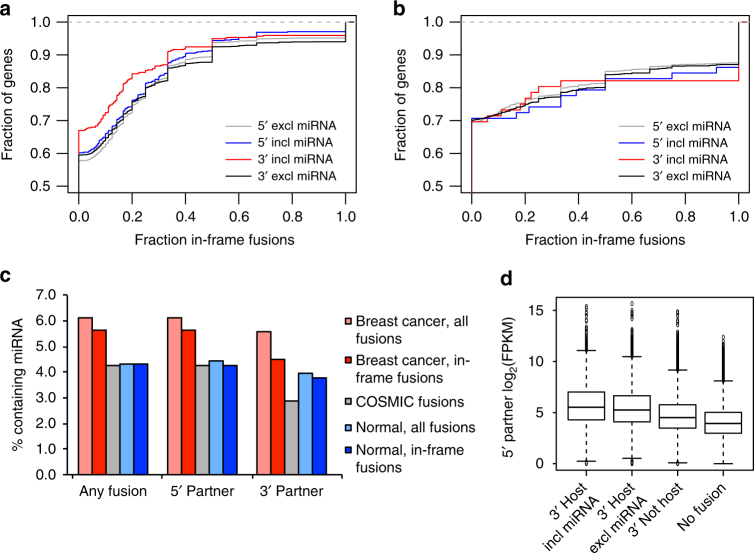
Fig. 2.
Fusion transcripts involving miRNA host genes as 3ʹ partners have fewer in-frame fusions. Cumulative distribution of the fraction of in-frame fusions among all fusion transcripts, plotted separately for 5ʹ and 3ʹ partner genes that lack or include miRNAs within the fused gene segments in our breast cancer data (a) and among fusion transcripts from approximately 30 different normal tissue types from ref. 27 (b) Percentage of genes with fusions that include miRNAs when analyzing all fusion types or only in-frame fusion transcripts for all protein-coding genes involved in fusions or among 5ʹ and 3ʹ partners separately, and, for comparison, among manually curated fusions in the COSMIC database (c). The 5ʹ partners of miRNA host genes have higher average expression than those of non-host genes, especially when the miRNA is included in the fused segment of the 3ʹ partner (d). Average expression was calculated separately for each 5ʹ partner gene among tumors with a fusion transcript matching either of the three 3ʹ partner categories. No fusion represents expression for the full set of 5ʹ partner genes in samples without fusion transcripts involving them. Expression is in log2(FPKM) (fragments per kilobase of exon model and million reads)