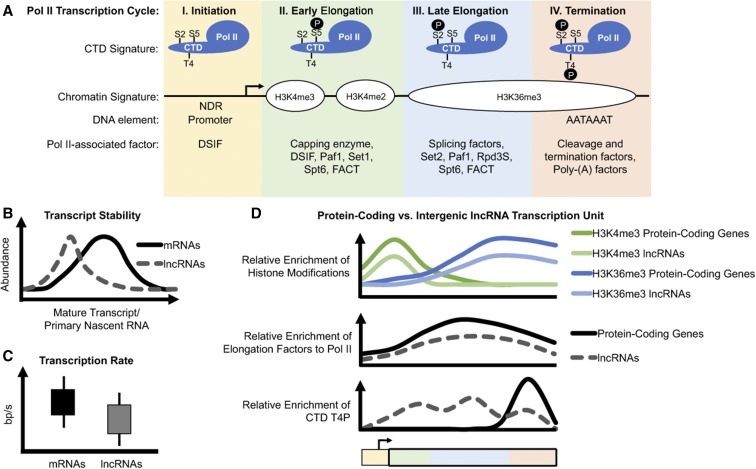
Figure 1.
Emerging differences in RNAPII behaviors at protein-coding and lncRNA transcription units. (A) The RNAPII transcription cycle at protein-coding genes is defined by four distinct stages. These stages are characterized by the association of RNAPII with transcription elongation factors and RNA processing factors, many of which are recruited by different post-translational modifications to the RNAPII CTD and/or histones. Only some of these factors are listed here. Active promoters are NDRs, while terminators are generally enriched for the AATAAAT consensus motif. (B) In general, lncRNAs are much less abundant than mRNAs, as lncRNAs are predominantly localized in chromatin where they are rapidly degraded (Schlackow et al. 2017). (C) Metabolic labeling studies using 4sU or 4tU reveal that RNAPII transcription rates are on average slower for lncRNAs (Barrass et al. 2015; Eser et al. 2016; Mukherjee et al. 2017). (D) Many lncRNAs also exhibit reduced levels of transcription-coupled histone marks such as H3K4me3 and H3K36me3 (Sun et al. 2015), decreased recruitment of many common elongation factors (Battaglia et al. 2017; Fischl et al. 2017), inefficient processing (Eser et al. 2016; Mukherjee et al. 2017; Schlackow et al. 2017), and early termination (Milligan et al. 2016; Schlackow et al. 2017). 4sU, 4-thiouridine; 4tU, 4-thiouracil; CTD, C-terminal domain; H3K, histone H3 lysine; lncRNA, long noncoding RNA; NDR, nucleosome-depleted regions; RNAPII, RNA Polymerase II.