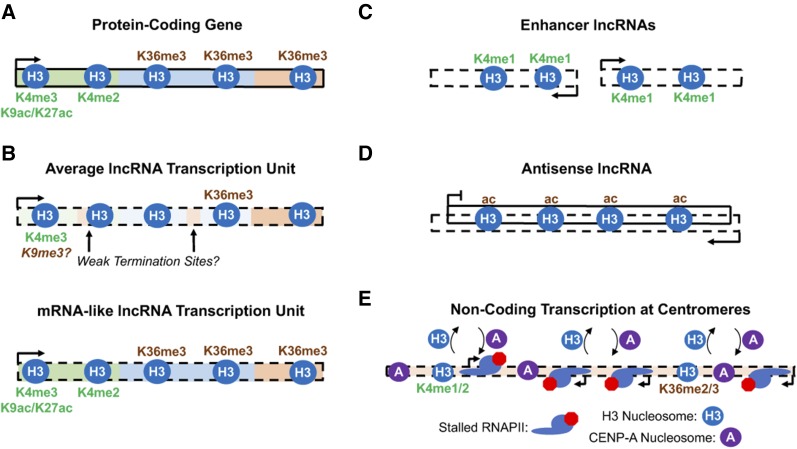
Figure 2.
Chromatin signatures of coding and noncoding transcription. (A) Distribution of transcription-coupled histone modifications across active protein-coding genes. H3K4me3 and H3 acetylation at K9 and K27 are enriched at promoters while H3K4me2 and H3K36me3 accumulate over gene bodies (Corden 2013). (B) Intergenic lncRNAs on average display reduced levels of H3K4me3 and H3K36me3 (Sun et al. 2015) and exhibit premature termination and defective nascent transcript processing (Schlackow et al. 2017). H3K9me3, a mark generally characteristic of repressive heterochromatin, has been detected at active lncRNA promoters in human cells (Méle et al. 2017). Importantly, some lncRNA transcription units also have more mRNA-like patterns of CTD and histone modifications. (C) Chromatin marks typically associated with active promoters (H3K4me3 and H3 acetylation) are depleted from noncoding transcription units at enhancer elements in higher eukaryotes. Instead, enhancers are enriched in H3K4me1 (Li et al. 2016). (D) lncRNA transcription antisense to protein-coding genes has been observed to correspond with reduced H3K36me3 and other marks commonly associated with active transcription elongation and instead enriched for H3 acetylation (Murray et al. 2015). (E) Noncoding transcription in centromeres has been observed to correspond with an absence of H3K4me3 and H3 acetylation. Instead, centromeric nucleosomes containing canonical H3 are enriched in H3K4me1/2 and H3K36me2/3 (Sullivan and Karpen 2004; Bergmann et al. 2011). Frequent transcription stalling in fission yeast centromeres has also been found to stimulate the incorporation of nucleosomes containing the H3-variant CENP-A, which epigenetically specifies centromeres (Catania et al. 2015). CENP-A, centromere protein-A; CTD, C-terminal domain; H3K, histone H3 lysine; lncRNA, long noncoding RNA; RNAPII, RNA Polymerase II.