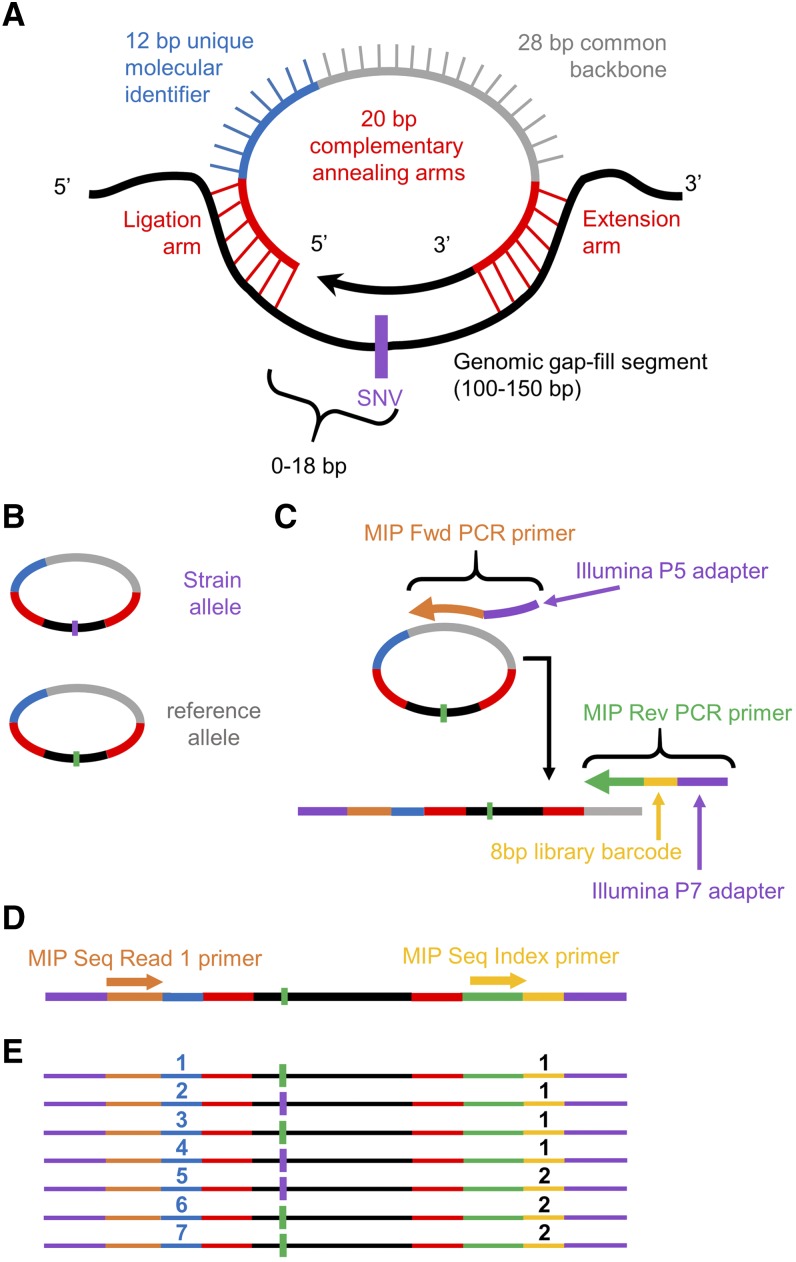
Figure 1.
MIPs design and workflow. The design of the smMIPs (Hiatt et al. 2013) was modified to relocate the unique molecular identifier. Overall, the MIP design incorporates two 20-bp annealing arms, a 12-bp molecular tag, and a 28-bp common backbone. After annealing to a target segment ranging in size from 100–150 bp (A), the MIP is gap-filled with high-fidelity polymerase capturing the sequences of interest and circularized via ligation (B). The SNV of interest is located within an 18-bp gap-fill window upstream of the ligation arm. Uncircularized DNA is then degraded via exonucleases. The remaining MIPs are linearized by combining with sequencing adapters and library barcodes during PCR amplification (C). A single-end read captures both the molecular tag, 5′-annealing sequence, and enough genomic sequence to confirm correct target capture (D). This information is then demultiplexed (E) and used as a means to compare sequence variant ratios. Fwd, forward; MIPs, molecular inversion probes; Rev, reverse; Seq, sequencing; smMIP, single-molecule MIP; SNV, single-nucleotide variant.