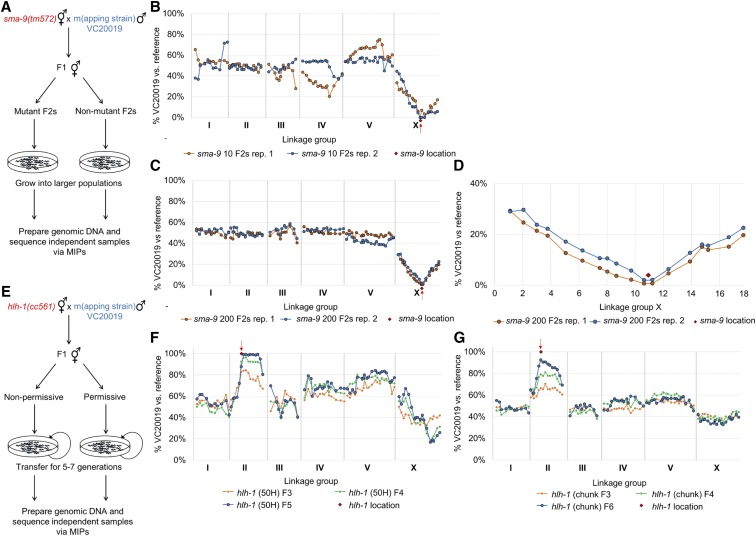
Figure 2.
Mapping of sma-9 and hlh-1 via VC20019 and MIP-MAP sequencing. Bulk segregant mapping scheme for sma-9 (A) using MIPs that target a number of VC20019-specific SNVs across the genome. Two replicate MIP-MAP samples of populations grown from 10 Sma phenotype F2 animals sufficiently identified a sma-9-associated interval on X within a 1.5–4.2 Mbp window (B) and replicates using 200 Sma F2 recombinants produced a 2.6 Mbp window (C). Closer examination of the MIP target sites on LGX show sma-9 is located equidistantly between two probes only 590 kbp apart (D). The hlh-1(cc561) temperature-sensitive embryonic lethal allele was mapped using a competitive fitness mapping method (E). PD4605 hermaphrodites were mated with VC20019 males and nonpermissive temperatures were used to select against the subsequent hlh-1(cc561) homozygous progeny. Starting with F1 cross-progeny and transferring mixed-stage subpopulations of animals (F and G) for up to seven generations, a region corresponding to the hlh-1 locus on LGII was successfully identified by the fixation of VC20019-specific SNVs. A mapping interval of 2.8–7.8 Mbp was identified by the F5 generation when transferring populations of 50 L1s (F). A 2.7-Mb interval was observed by the F6 generation when transferring 4-cm2 chunks of mixed-stage animals (G). Each line present in (F and G) represents mapping data from a different generation for the same biological replicate. MIP, molecular inversion probe; MIP-MAP, MIP mapping; SNV, single-nucleotide variant.