Figure 1.
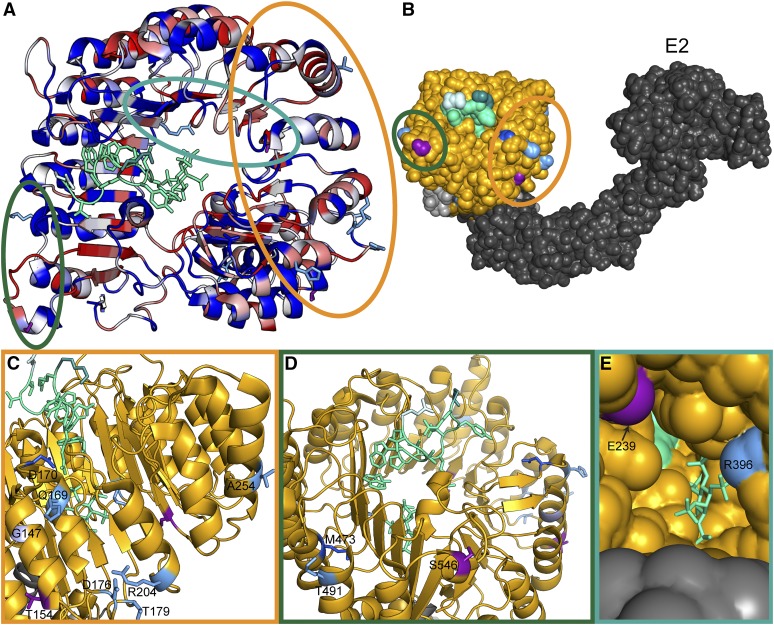
Clusters of natural variation in TIR1 and AFB2. (A) Identified nonsynonymous polymorphisms tend to occur in residues of high diversity within the Arabidopsis AFB family. A top down view of the LRR domain of the TIR1 structure [Protein Data Bank (PDB): 2P1Q] is shown with the F-box domain in the bottom right and the LRR domain spiraling counterclockwise. The backbone of the TIR1 structure (Tan et al. 2007) was colored according to protein sequence diversity with conserved residues in blue and diverging residues in red. Diversity was calculated as Shannon entropy using an alignment of the protein sequences of the Arabidopsis AFB family (TIR1, AFB1–5). All nonsynonymous polymorphisms are shown as sticks. AFB2 variants are in light blue and TIR1 variants are in purple. Previously identified TIR1 mutations are in dark blue (Ruegger et al. 1998; Yu et al. 2013). The IAA7 degron is shown as a light green ribbon with sidechains as sticks. The N-terminal residue of the IAA7 degron is in lighter green and the C-terminal residue is darker green. Circles around polymorphisms match the detailed views shown in C–E. (B) Polymorphisms face the Cullin subunit of the predicted SCFTIR1 structure. ASK1 (light gray) was aligned with SKP1 from the human SKP2-SKP1-Cul1-RBX1 structure (PDB: 1LDK, shown in dark gray), docking with TIR1 (gold). Putative E2 location is labeled. (C) The dimerization domain on the N-terminal side of the LRR horseshoe contains the majority of natural variation in TIR1 and AFB2. The tir1-1 allele (tir1G147D) is in light purple. (D) Two variants were located on the C-terminal side of the LRR close to the N terminus of the degron. (E) Two additional variants were located inside the LRR horseshoe, near the inositol–hexakisphosphate cofactor.