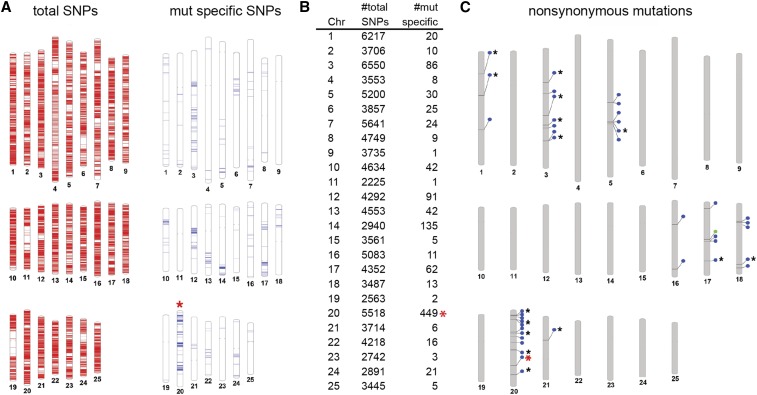
Figure 2.
Mapping and candidate gene identification in dmh35. To identify mutant specific haplotypes, first all SNPs in the genome are identified. After exclusion of all SNPs that are present in the sibling pool, only mutant specific SNPs are left. The highest number of mutant specific SNPs on a chromosome should indicate the chromosome carrying the mutation. (A) The red bars in the diagram on the left represent SNPs identified in the mutant and sibling pool on each of the 25 chromosomes. Blue bars in the diagram on the right represent mutant-specific SNPs on each of the 25 chromosomes. (B) Summary of the number of total and mutant-specific SNPs per chromosome. (C) In a second step, all nonsynonymous changes are identified in the genome. Blue circles indicate missense mutations; green circles mark nonsense mutations; black asterisks mark mutations predicted to be deleterious; the red asterisk indicates the position of the G138D missense mutation in connexin 43, shown to be linked to the dmh35 mutant phenotype.